Abstract
Competing fungi in white button mushroom (Agaricus bisporus, champignon) cultivation causes significant episodic losses. One of these competing fungi is known as “smoky mould”. It owes its name to the production of high numbers of spores after the disturbance of compost, which resembles smoke. We investigated strains isolated from smoky mould cases in the Netherlands, UK and Canada and show that these outbreaks were caused by a new Penicillium species, named P. hermansii sp. nov. (type strain CBS 124296T). Several Penicillium species are reported to cause smoky mould. However, we so far have no indications that smoky mould is caused by other Penicillium species than P. hermansii. This species belongs to section Exilicaulis and differs from other Penicillia by its slow growth rate on Czapek yeast agar (CYA) and malt extract agar (MEA) and its inability to grow on CYA supplemented with 5% salt and CYA and MEA incubated at 37 °C.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A critical step during mushroom production is the colonisation of the pasteurised phase II compost by Agaricus bisporus. Competitive moulds from spawn-run compost can have devastating effects on production levels. An important and well-known example is Trichoderma aggressivum and much research is performed on this species (Kosanovic et al. 2015; O’Brien et al. 2014; O’Brien et al. 2017; Radvanyi et al. 2016). Another less commonly and poorly documented competing contaminant is known as ‘smoky mould’, which is capable of wiping out a complete crop (Fletcher and Gaze 2008; Grogan and Harvey 1999; Grogan et al. 2000). In the last decade, several outbreaks occurred in the Netherlands (C. Hermans, pers. comm.). The first signal of infection by smoky mould is an increase of temperature of the compost during growth that is difficult to control even by lowering air temperature. Mushrooms grown on compost with a minor smoky mould infection are slightly paler, mature quicker and there is usually reduced pinning. In more severely infected compost, areas lacking growth and even complete empty mushroom beds occur. Digging into these infested areas releases large clouds of spores resembling smoke, hence the name smoky mould.
Various names, such as Penicillium chermesinum, P. citreonigrum, P. implicatum and P. fellutanum have been applied to smoky mould outbreaks (Baars et al. 2011; Beyer 2002; Fletcher and Gaze 2008; Grogan et al. 2001). These species are distantly related to each other and belong to different Penicillium sections (Houbraken and Samson 2011; Visagie et al. 2014). The use of these names in literature might suggest that smoky mould is caused by multiple species. However, in the past Penicillium, identification was primary based on phenotypic and physiological characters. It is probable that only one fungus is involved and that some or all of these identifications are incorrect.
In this study, strains isolated from smoky mould outbreaks in the Netherlands, Canada and UK were studied using a polyphasic approach. Physiological, macro- and microscopical characteristics combined with partial calmodulin (CaM), β-tubulin (BenA) and RNA polymerase II second largest subunit (regions 5–7) (RPB2) sequences demonstrate that this set of strains represents a new species in Penicillium section Exilicaulis.
Material and methods
Strains
An overview of examined strains is presented in Table 1. The 16 investigated strains were isolated from compost or the air of mushroom production farms, where Agaricus bisporus cultivations were infected with smoky mould. The strains that were isolated in different years were from different outbreaks and different production farms. No information is recorded at farm level and strains isolated in the same year could be obtained from more than one production location. Two of the strains are legacy strains from previous outbreaks, HRI 1043-D from the UK and the main strain mentioned in previous literature (Fletcher et al. 1989; Grogan et al. 2000), and VM-2 from Canada.
Phenotypic and physiologic characterisation
All pure cultured strains were inoculated at three points on Czapek yeast autolysate agar (CYA), CYA supplemented with 5% NaCl (CYAS), malt extract agar (MEA, Oxoid), oatmeal agar (OA), creatine sucrose agar (CREA), dichloran 18% glycerol agar (DG18), yeast extract sucrose agar (YES) (Frisvad and Samson 2004). Additional CYA and MEA plates were incubated at 15, 30 and 37 °C. Colony diameters and other macroscopic colony characteristics (e.g. colour conidia, colony texture) were recorded after 7 days of incubation at 25 °C. Mounts were made from the MEA medium in lactic acid and microscopic features were studied by light microscopy using a Zeiss Axiokop 2 Plus. The temperature-growth response was investigated on CYA by recording diameters of three-point inoculated colonies of seven strains (CBS 122432, CBS 124296, CBS 124297, DTO 032-A2, DTO 032-A3, DTO 173-C9 and DTO 173-D1) after 14 days at 6–36 at 3 °C intervals and 40 °C. Alphanumeric codes for conidial and reverse colours refer to (Kornerup and Wanscher 1978). Isolates were also examined for the production of alkaloids with Ehrlich reagent using the filter paper method described by Lund (1995). The growth rate of two P. hermansii strains (CBS 124296 and DTO 173-D4) and four other Penicillium species (P. brevicompactum DTO 099-D1; P. chrysogenum CBS 306.48; P. daleae DTO 099-B8; P. glabrum, DTO 099-A6) was studied on an autoclaved compost based medium containing 250 g Agaricus grown compost (phase III) and 15 g agar per litre.
DNA extraction, sequencing and phylogenetic analysis
Strains were grown for 7–14 days at 25 °C on MEA prior to DNA extraction. Genomic DNA was extracted using the Ultraclean Microbial DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, USA) following the manufacturer’s protocol. The ITS barcode and parts of the BenA, CaM and RPB2 genes were amplified, sequenced and annotated according to the method described by Houbraken and Samson (2011) and Houbraken et al. (2012). Newly generated sequences are deposited in GenBank with accession numbers MG386210–MG386247 and MG333469–MG333479.
Publically available BenA, CaM and RPB2 sequences of the type strains of species belonging to section Exilicaulis were retrieved from GenBank and supplemented with the newly generated sequences. A concatenated dataset with BenA, CaM and RPB2 sequences including nine smoky mould strains was generated and used to determine the phylogenetic relationship of the smoky mould strains with other members of Penicillium section Exilicaulis. Single gene phylogenies were made by combining the generated sequences from smoky mould isolates with sequences of strains belonging to the P. parvum-clade (Visagie et al. 2016) of sect. Exilicaulis. The sequence data sets were aligned using MAFFT (Katoh and Standley 2013). Prior to analysis, the optimal substitution model was determined in MEGA v.6.06. (Tamura et al. 2013), utilising the Akaike information criterion (AIC). Maximum likelihood (ML) analysis of the single gene datasets were analysed in MEGA 6.06 and the combined dataset in RAxML-VI-HPC v. 7.0.3 (Stamatakis 2006) using the GTRGAMMA substitution model. The robustness of the phylograms was evaluated by 1000 bootstrap (bs) runs. Bayesian inference (BI) was performed in MrBayes v.3.2.1 (Ronquist et al. 2012) using Markov chain Monte Carlo (MCMC) algorithm. The analysis stopped when the average standard deviation of split frequencies reached 0.01. The sample frequency was set to 100; the first 25% of trees were removed as burnin. Convergence and ESS values of the runs were examined by Tracer 1.6 (Rambaut et al. 2014).
Results
Phylogeny
The total length of the combined data set was 1729 characters (individual datasets: BenA 421; CaM, 518; RPB2, 788 characters) and the proportion of gaps was 5.7%. The phylogram was drawn to scale and the results of this analysis are shown in Fig. 1. Nine smoky mould isolates were included in this analysis and those isolates form a unique clade within section Exilicaulis. Two well-supported clades are present in this section and the smoky mould strains group in a cluster previously named the Penicillium parvum-clade (Visagie et al. 2016). Penicillium canis and P. striatisporum are basal to the clade containing the smoky mould isolates; however, statistical support is lacking. With the data available, it was not possible to conclusively identify the phylogenetic sister species of the smoky mould isolates.
Best-scoring maximum likelihood tree using RAxML based on a combined dataset with partial BenA, CaM and RPB2 sequences, showing the relationship of species accommodated in section Exilicaulis. The phylogram is rooted with P. glabrum CBS 125543. Bootstrap percentages and posterior probability values are presented at the nodes. Values less than 70% bs or 0.95 pp are not shown and branches with more than 95% bs and 1.00 pp are thickened. The bar indicates the number of substitutions per site
BenA, CaM and RPB2 gene sequences were generated to confirm the phylogenetic coherence of the smoky mould strains at the species level, and their relationship with other species in the P. parvum-clade. The Tamura Nei (TN93) model using the gamma distribution (+G) was found to be optimal for the BenA data set (length 372 sites); and the TN93 model using a discrete gamma distribution with invariant sites (G+I) was optimal for the CaM (length 518 sites) and RPB2 (length 789 sites) datasets. The best-scoring ML trees are presented in Fig. 2 and show that the smoky mould strains form a coherent species level clade, distinct from other members of the P. parvum-clade. The smoky mould strains cluster together in trees based on all loci on a strongly supported branch (> 95% bs, 1.00 pp). Most bootstrap percentages and posterior probability (pp) values were low (< 70% bs, < 0.95 pp) in these gene trees.
Maximum likelihood (ML) trees generated for BenA, CaM and RPB2 with MEGA6. The phylograms are rooted with P. corylophilum CBS 312.48. Bootstrap percentages and posterior probability values are presented at the nodes. Values less than 70% bs or 0.95 pp are not shown and branches with more than 95% bs and 1.00 pp are thickened. The bar indicates the number of substitutions per site
Morphology and physiology
Phenotypic characters of the smoky mould strains were recorded and compared with each other. All strains displayed similar macro- and microscopical characters. Most strikingly, the isolates grew slow on CYA and MEA, even at an optimum growth temperature (27–30 °C). Furthermore, the conidiophores had short stipes (10–40 (− 120) μm) and the strains did not grow on CYAS, and CYA and MEA incubated at 37 °C. The optimum growth temperature on CYA was between 27 and 30 °C, with a colony diameter of 15–18 mm after 14 days. The minimum growth temperature was between 12 and 15 °C and the maximum growth temperature between 33 and 36 °C. Strains incubated at 33 °C reached 11–14 mm and no growth was observed at 36 °C. The colony diameters of the smoky mould strains on autoclaved compost agar (CA) were similar to those on MEA (CA 9–12 vs MEA 9–12 mm). Also the growth density was similar on both media. The other examined Penicillium species grew well on CA and their colony diameters were similar or slightly smaller than on MEA (P. brevicompactum, CA 18–22 vs MEA 20–24 mm; P. chrysogenum, CA 40–43 vs MEA 30–35 mm; P. daleae, CA 24–27 vs MEA 34–37 mm and P. glabrum, CA 38–40 vs MEA 41–45 mm).
Taxonomy
Based on phylogenetic coherence of the smoky mould causing isolates and their unique phenotypic and physiologic characters, we describe them here as a new species named Penicillium hermansii sp. nov.
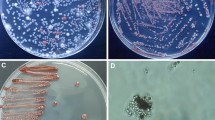
Penicillium hermansii Houbraken, Seifert & Samson, sp. nov. Mycobank MB823949 (Fig. 3).
In: Penicillium subgenus Aspergilloides section Exilicaulis.
Etymology: Named after Con Hermans, who studied the occurrence of smoky mould and other competing fungi in the Netherlands.
Diagnosis: Slow growth on CYA (5–10 mm) and MEA (8–15 mm) at 25 °C after 7 days incubation; no growth on CYAS; no growth on MEA and CYA incubated at 37 °C; conidiophores biverticillate and short-stiped, 10–40 (−120) μm.
Typus: the Netherlands, ex compost with Agaricus bisporus, 2008, collected by C. Hermans, isolated by J. Houbraken (holotype CBS H-21028, culture ex-type CBS 124296 = DTO 079-D5).
ITS barcode: ITS = MG333472 (alternative markers: BenA = MG386214, CaM = MG386229, RPB2 = MG386242). All examined P. hermansii strains share identical ITS, BenA, CaM and RPB2 sequences. The species can be differentiated from other known Penicillium species by ITS sequences.
Distribution and ecology: The Netherlands, United Kingdom, Canada. Isolated from compost colonised by Agaricus bisporus and from the air of white button mushroom production farms.
Colony diameter: 7 days, in mm, 25 °C (unless stated otherwise): CYA 5–10; CYA15°C no growth or spore germination; CYA 30 °C 7–12; CYA 37 °C: no growth; MEA 8–15; YES 5–9; DG18 3–7; CYAS no growth; OA 10–15; creatine agar no growth or spore germination. Optimum growth temperature on CYA 27–30 °C.
Colony characters: Sporulation on CYA absent to moderate; colonies entire, plane, velvety; mycelium white, sporulation grey-green to greyish-brown green (~ 5D3); exudate absent; soluble pigments absent; colony reverse brown (5C3–E3). Soluble pigments on YES absent, sporulation light, (pale) grey-green or inconspicuous; exudate absent, reverse brown. Colonies on MEA velvety, dull green or grey-green (25–26E3), reverse brown (5D3). Ehrlich reaction negative.
Micromorphology: Conidiophores borne from surface; slightly vesiculate; stipes short, 10–40 (− 120) × 2–3.5 μm, with walls smooth or very finely roughened; conidiophores of freshly isolated strains bearing terminal verticils of 2–4 metulae, and sometimes subterminal or intercalary metulae present; in degenerated strains monoverticillate conidiophores predominantly present. Terminal and subterminal metulae unequal in length, 10–20 (− 25) × 2.0–3.5 μm, often with inflated apex, 3–5 μm wide. Phialides ampulliform, in verticils of 4–10, closely packed, 7–9 × 2–3.5 μm. Conidia globose to subglobose, smooth-walled, 2.0–2.5 μm. Ascomata and sclerotia not observed.
Discussion
Grogan et al. (2001) expressed uncertainty about the identity of the smoky mould fungus in mushroom cultivation because taxonomic authorities that they consulted did not agree on a common identification. Most identifications of smoky mould isolates were performed before the era of DNA sequence-based identification. Grogan et al. (2000) studied the identity of four smoky mould isolates (including isolate HRI 1043-D) using ITS sequences, but the exact identification of the species involved in smoky mould problems remained uncertain. As demonstrated in this paper, the smoky mould isolates also turn out be a previously undescribed species. Although several Penicillium species (e.g. P. chermesinum, P. citreonigrum, P. implicatum and P. fellutanum) were named as causal agents of smoky mould, we suggest that these outbreaks were probably all caused by P. hermansii alone. Strains referred to as smoky mould in the literature share a slow growth rate on agar media and most were reported to be monoverticillate (Baars et al. 2011; Beyer 2002; Fletcher and Gaze 2008). Penicillium hermansii also grows slowly, but fresh isolates have terminal verticils of 2–4 metulae.
Phylogenetically, P. hermansii is most closely related to P. canis, P. catenatum, P. erubescens, P. guttulosum, P. menonorum, P. nepalense, P. papuanum, P. parvum, P. pimiteouiense, P. rubidurum, P. striatisporum and P. vinaceum that all align to the P. parvum-clade of section Exilicaulis. These species share a slow growth rate on CYA and MEA and produce short-stiped, monoverticillate conidiophores. Like the other members of this clade, P. hermansii also grows slowly on agar media and along with P. nepalense and P. catenatum is among the slowest growing species of the clade. Morphologically, it differs from all other species of this clade by the production of biverticillate conidiophores, with monoverticillate conidiophores only seen in older, presumably degenerated strains. Penicillium hermansii also differs from most of the phylogenetically related species by its inability to grow at 37 °C, a character shared only with P. nepalense (Pitt 1980 [‘1979’]; Takada and Udagawa 1983). Using the dichotomous key published in the Penicillium monograph of Pitt (1980 [‘1979’]), P. hermansii keys out as P. fellutanum. That species most obviously differs from P. hermansii by its faster growth rate on CYA (5–10 vs 17–25 mm).
In addition to its distinctive phenotype, P. hermansii is also unique in Penicillium for its association with Agaricus-colonised compost. Until now, this species has never been isolated from other habitats, despite extensive surveys of soil, food, indoor air and other substrates from all over the world. Strong associations between Penicillium species and particular habitats have been known for a long time (Westerdijk 1949). For example, P. italicum, P. ulaiense and P. digitatum are associated with rot of citrus fruits and P. allii is strongly associated with rot of garlic. Generally, Penicillium species with a specific association to certain substrates grow well on standard laboratory media and do not require special compounds from its associated natural source, e.g. citrus peels or garlic (Frisvad and Samson 2004). In contrast, P. hermansii grows slowly on general purpose agar media CYA, MEA and DG18, but observations in practice show that the species thrives well on Agaricus grown compost. All Penicillium strains used in this study grew well on CA. The components of this autoclaved medium cannot explain the profuse growth of P. hermansii in Agaricus colonised compost. The compost composition might therefore not be the defining parameter for growth of P. hermansii. Actually, many Penicillium species occur in compost and seem to have little or no effect on Agaricus growth (Grogan et al. 2001).
Very little work has been carried out on competition with Agaricus by P. hermansii and most research on competing fungi in Agaricus production has focused on Trichoderma aggressivum. This species grows quickly on agar media and seems to compete with Agaricus for space and nutrients effectively. The association of T. aggressivum with Agaricus production is probably first based on chemical communication via extrolites, including volatiles, and/or extracellular enzymes. For example, the mycelium of A. bisporus is required for induction of intensive sporulation in T. aggressivum (Krupke et al. 2003; Mamoun et al. 2000; Mumpuni et al. 1998; Seaby 1987; Seaby 1996). Similarly, Beyer (2002) indicated an interaction between the mycelium of Agaricus and smoky mould and suggested that the Agaricus is either parasitised or repressed. More research is needed to understand the biology of smoky mould in button mushroom cultivation.
References
Baars J, Rutjens J, Mumm R (2011) Can volatiles emitted by compost during spawn run be used to detect green mould infection early? In: Savoie J-M, Foulongne-Oriol M, Largeteau M, Barroso, G. (eds) Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products (ICMBMP7), vol 1. INRA, Villenave d’Ornon Cedex, pp 474–483
Beyer DM (2002) Pest species biology and control—weed and indicator moulds. In: Mushroom integrated pest management handbook. Pennsylvania State University Press, Pennsylvania, pp 61–74
Fletcher JT, Gaze RH (2008) Mushroom pest and disease control, a colour handbook. Academic Press, San Diego
Fletcher JT, White PP, Gaze RH (1989) Mushrooms—pests and diseases. 2. Intercept, Andover
Frisvad JC, Samson RA (2004) Polyphasic taxonomy of Penicillium subgenus Penicillium—a guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol:1–173
Grogan H, Harvey L (1999) The effects of compost moulds in mushroom compost. Horticulture Research International, Warwick
Grogan HM, Scruby A, Harvey L (2000) Moulds in spawn-run compost and their effect on mushroom production. Science and Cultivation of Edible Fungi, Vols 1 and 2:609–615
Grogan H, Parker L, Scruby A (2001) Survey of compost moulds in traditional and bulk Phase III (spawn-run) compost. Horticulture Research International, Warwick
Houbraken J, Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70:1–51
Houbraken J, Spierenburg H, Frisvad JC (2012) Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 101:403–421
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kornerup A, Wanscher JH (1978) Methuen handbook of colour, 3rd edn. Eyre Methuen Ltd., London
Kosanovic D, Potocnik I, Vukojevic J, Stajic M, Rekanovic E, Stepanovic M, Todorovic B (2015) Fungicide sensitivity of Trichoderma spp. from Agaricus bisporus farms in Serbia. J Environ Sci Heal B 50:607–613
Krupke OA, Castle AJ, Rinker DL (2003) The North American mushroom competitor, Trichoderma aggressivum f. aggressivum, produces antifungal compounds in mushroom compost that inhibit mycelial growth of the commercial mushroom Agaricus bisporus. Mycol Res 107:1467–1475
Lund F (1995) Differentiating Penicillium species by detection of indole metabolites using a filter-paper method. Lett Appl Microbiol 20:228–231
Mamoun ML, Savoie J-M, Olivier J-M (2000) Interactions between the pathogen Trichoderma harzianum Th2 and Agaricus bisporus in mushroom compost. Mycologia 92:233–240
Mumpuni A, Sharma HSS, Brown AE (1998) Effect of metabolites produced by Trichoderma harzianum biotypes and Agaricus bisporus on their respective growth radii in culture. Appl Environ Microbiol 64:5053–5056
O’Brien M, Grogan H, Kavanagh K (2014) Proteomic response of Trichoderma aggressivum f. europaeum to Agaricus bisporus tissue and mushroom compost. Fungal biology 118:785–791
O’Brien M, Kavanagh K, Grogan H (2017) Detection of Trichoderma aggressivum in bulk phase III substrate and the effect of T. aggressivum inoculum, supplementation and substrate-mixing on Agaricus bisporus yields. Eur J Plant Pathol 147:199–209
Pitt JI (1980 [1979]) The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, London
Radvanyi D, Gere A, Sipos L, Kovacs S, Jokai Z, Fodor P (2016) Discrimination of mushroom disease-related mould species based solely on unprocessed chromatograms. J Chemom 30:197–202. https://doi.org/10.1002/cem.2777
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer, version 1.6. http://beast.bio.ed.ac.uk/Tracer
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Seaby DA (1987) Infection of mushroom compost by Trichoderma species. Mushroom Journal 179:355–361
Seaby DA (1996) Differentiation of Trichoderma taxa associated with mushroom production. Plant Pathol 45:905–912
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Takada M, Udagawa SI (1983) Two new species of Eupenicillium from Nepalese soil. T Mycol Soc Jpn 24:143–150
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Visagie CM et al (2014) Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371
Visagie CM, Seifert KA, Houbraken J, Samson RA, Jacobs K (2016) A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7:75–117
Westerdijk J (1949) The concept ‘association’ in mycology. Antonie Van Leeuwenhoek 15:187–189
Acknowledgements
We thank Nancy Nickerson, Robert Davies and Leonard North for supplying the Canadian isolate of P. hermansii and Gerry Louis-Seize for early phylogenetic studies of that strain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Hans-Josef Schroers and Marc Stadler
This article is part of the “Special Issue on hyphomycete taxonomy and diversity in honour of Walter Gams who passed away in April 2017”.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Houbraken, J., Seifert, K.A. & Samson, R.A. Penicillium hermansii, a new species causing smoky mould in white button mushroom production. Mycol Progress 18, 229–236 (2019). https://doi.org/10.1007/s11557-018-1407-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-018-1407-4