Abstract
Automated nucleus segmentation is considered the gold standard for diagnosing some severe diseases. Accurate instance segmentation of nuclei is still very challenging because of the large number of clustered nuclei, and the different appearance of nuclei for different tissue types. In this paper, a neural network is proposed for fast and accurate instance segmentation of nuclei in histopathology images. The network is inspired by the Unet and residual nets. The main contribution of the proposed model is enhancing the classification accuracy of nuclear boundaries by moderately preserving the spatial features by relatively d the size of feature maps. Then, a proposed 2D convolution layer is used instead of the conventional 3D convolution layer, the core of CNN-based architectures, where the feature maps are first compacted before being convolved by 2D kernel filters. This significantly reduces the processing time and avoids the out of memory problem of the GPU. Also, more features are extracted when getting deeper into the network without degrading the spatial features dramatically. Hence, the number of layers, required to compensate the loss of spatial features, is reduced that also reduces the processing time. The proposed approach is applied to two multi-organ datasets and evaluated by the Aggregated Jaccard Index (AJI), F1-score and the number of frames per second. Also, the formula of AJI is modified to reflect the object- and pixel-level errors more accurately. The proposed model is compared to some state-of-the-art architectures, and it shows better performance in terms of the segmentation speed and accuracy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Detection and segmentation of nuclei in histology images are considered crucial processes for diagnosing, grading and even for prognosis prediction of many diseases, such as most cancer types and Alzheimer [1,2,3,4]. In current clinical practices, the examination of hematoxylin and eosin (H&E)-stained tissue images (to analyze nuclei density, morphology and shape) is carried out manually, by means of pathologists. However, the manual analysis results in many problems such as inter- and intra- observer variability, inability to assess fine visual features and a huge amount of time to examine Whole Slide Images (WSI) [5, 6]. With the great revolution in computer vision and image processing techniques, many manual assessment problems of histology images have been addressed [7,8,9]. Several traditional image processing methods for automatic nucleus detection and segmentation were presented, such as Otsu thresholding [10], Marker-controlled watershed segmentation [11] and other region growing, morphology, feature extraction and color-based thresholding operations [12,13,14]. However, such methods are mainly based on predefined colors, shapes, or textures of nuclei that cannot be constant for all cases, such as different types of tissues, different grading of disease or the wide spectrum of tissue morphologies. In addition, due to noise and various staining concentration that appear in H&E stained images, such traditional methods fail to produce accurate and robust nucleus segmentation. Furthermore, those approaches produce either under- or over-segmentation of clustered and overlapping nuclei [15]. Throughout the past few decades, learning-based nucleus detection and segmentation methods have been proposed, where handcrafted features are extracted, such as color histograms, texture, morphology, optical density, geometric and other characteristics of nuclei [16,17,18]. The extracted features are then fed into Machine Learning (ML)-based models, such as Random Decision Forest (RDF) [19], the K-Nearest Neighbors (KNN) [20] and Support Vector Machine (SVM) [21] algorithms to produce the nuclear probability map within the histology images. However, ML approaches depend on predefined features, and the parameters are adapted based on trial-and-error during training in order to achieve the required performance. This makes it difficult to consider them as generalized nucleus segmentation approaches. Recently, many Deep Learning (DL) models have been proposed for nuclear detection and segmentation that address the limitations of the handcraft feature-based models, where most of them are trained, based on pre-annotated datasets, including nuclear and non-nuclear pixels. This enables models to extract more detailed, sophisticated and hidden features that cannot be easily recognized by traditional and standard handcraft feature-based approaches, to achieve a generalized robust nucleus segmentation [22]. Deep learning-based segmentation approaches are divided into semantic segmentation and instance segmentation. In semantic segmentation, the main concern is only distinguishing nuclear from non-nuclear pixels in the histology image. On the other side, instance segmentation is concerned with distinguishing and segmenting individual nuclei (objects), which is very important to help pathologists study the morphology, size and density of various nuclei in the same tissue to achieve accurate diagnosis, grading and prognosis [23,24,25,26]. Yet, the main challenge in most introduced instance nuclear segmentation approaches is separating overlapping and clustered nuclei. However, that is mostly at the expense of the computational complexity and processing time. Moreover, most of them are trained based on a certain type of nuclei, so they cannot be applied to various types of tissues.
In this work, a real-time architecture for accurate instance nuclear segmentation is proposed. The proposed architecture is inspired by the Unet [27] and residual nets [28]. The model was trained on multi types of nuclei of different kinds of tissues. The main contribution of the proposed model is that it moderately preserves the spatial features while reducing the computational complexity and processing time; minimizing the processing time is crucial since up to 1000 WSIs can be analyzed and diagnosed per day, in one clinical setup. The relative preservation of spatial features is important, especially, to identify the minor-class pixels that are the boundary pixels, generally, and the common pixels among touching and overlapping nuclei, specifically. To achieve that, the size of feature maps is not intensively reduced; instead, the channels of feature maps are compacted into one channel. Then, a proposed 2D convolution layer is used, instead of the conventional 3D convolution layer, which significantly reduces the processing time. Also, the rational preservation of spatial features enables reducing the number of layers, required to compensate the spatial feature loss throughout the model architecture. This has a considerable impact on the computational complexity and hence the training and inference time. Also, the segmentation accuracy of the boundary pixels, especially the ones between intersecting nuclei, are improved. The performance of the proposed model was evaluated by the Aggregated Jaccard Index (AJI), F1-score and the number of frames per second (FPS) that represents the number of segmented images by the proposed model per second. In addition to that, a modification was made to the AJI to make it more simulating to the pathologists’ way of thinking, so that, it reflects the real pixel-level mismatch, over- and under-segmentation cases, more accurately. Experimental results reveal that the proposed model provides better performance than some state-of-the-art approaches, whose architectures are complicated.
The remaining sections of the paper are organized as follows: Sect. 2 outlines related works. In Sect. 3, the methodology of the proposed model is explained, whereas the datasets, evaluation metrics, the modified AJI and implementation details are presented in Sect. 4. The time analysis of the proposed 2D convolution layer is discussed in Sect. 5. The results are demonstrated in Sect. 6, then the conclusion is drawn in Sect. 7.
2 Related work
Nuclei instance segmentation models are divided into box-based and box-free approaches. Most box-based instance segmentation models detect nuclei, using region proposals then segment the candidates to produce individual nuclear segmentation by classifying pixels inside the candidate ROI into background and foreground ones. Some standard DL approaches are used as a streamline in most box-based instance segmentation models such as the Single Shot Detector (SSD) [29], Fast- and Faster-R-CNN [30, 31], Mask R-CNN [32] and Retina Net [33]. Yi et al. [34] implemented a nuclei instance segmentation model, where the Unet architecture and the single shot multi-box detector were combined together. The attention mechanisms were used to improve the detection and segmentation accuracy, although they increase the computational complexity. In [35], the Mask R-CNN was used to, accurately, detect the candidate ROIs of nuclei. Hao Liang et al. [36] integrated the Guided Anchoring (GA) with the Region Proposal Network (RPN) to implement a GA-RPN module that generates candidate proposals for nuclei detection, then the Mask R-CNN was applied on the extracted ROI, for nuclear instance segmentation. A fast and accurate region-based nuclei instance segmentation algorithm was presented by Cheng et al. [37]. The architecture consists of detection and segmentation blocks. The bounding boxes of nuclei are detected by applying the feature pyramid network to combine both shallow and deep features. Accordingly, the feature maps are cropped and fed into the Unet architecture to improve the accuracy of instance segmentation. In [38], a context-refined neural network was presented to detect the ROIs of individual nuclei and eliminate the background. However, the system failed to detect small nuclei. In region-based instance segmentation models, the resultant accuracy mainly depends on the candidates of nuclear ROI that is affected by clustered and overlapping nuclei. On the other side, Box-free instance segmentation methods do not have to detect the ROI of nuclei, first, to segment them; instead, they apply pixel-wise segmentation to the whole image. In [25], a three-class CNN was used to produce a ternary map to segment inter-nuclear boundaries in the presence of clustered nuclei, then post-processing was done to, exactly, detect the nuclei contour. In [39], a dual Unet model was used, one for boundary detection and another for nuclei center detection, where two weight maps for nuclei center detection and boundary segmentation were proposed to enhance segmentation. In [40], the USE-Net architecture was introduced, where squeezing and excitation blocks were used to allow features recalibration by emphasizing the useful features and discarding the useless ones. The network also produces markers for nuclei to separate overlapping nuclei, by means of the watershed algorithm, and generate the final instance segmentation of nuclei. In [41], the authors introduced a cross-staining style method to synthesize nuclei pathology images, based on adversarial learning, to overcome the issue of insufficient large dataset for the training process. Then, a WNS-Net [42] was applied to segment the output of the synthesized branch, to detect the nuclei centers, without the need of annotating the whole nuclei or their boundaries. However, this can affect the segmentation result of overlapping nuclei. In [43], a Recursive DL (R-DL) strategy was introduced to train deep learning models, using incomplete annotations, and the Unet was used as a backbone for simultaneous detection and instance segmentation. The Unet was trained by a number of individual positive (nucleus) and negative (background) patches, selected from incompletely annotated pathologic dataset. However, the model was trained and tested based on a dataset that consists of only one type of nuclei, hence the model cannot support generalized nuclear segmentation. Also, the authors neglected the effect of intersecting nuclei that can affect the instance segmentation result. In [44], two independently trained Unet models were used for instance segmentation. The first Unet produces the semantic segmentation of nuclei, while the second one is used to predict the distance maps for all nuclei instances. To remove false local maxima, Gaussian smoothing filter was applied, then the derived local maxima were used as seed points of the watershed algorithm, whereas the semantic segmentation result of the first Unet model was used as a mask. In [23], the top-ranked technique was implemented using the Unet architecture, whose encoder was implemented using Resnet50 [28]. The network predicts two outputs, simultaneously, that represent the segmentation of nuclei and their contours. The instance segmentation is obtained by subtracting the predicted contour pixels from the predicted nuclei pixels. In [45], a Unet-based dual output neural network was introduced to predict the nuclei contours and inner pixels. The residual blocks and channel attention mechanisms were applied to increase the accuracy of instance segmentation. S. Graham et al. introduced a novel CNN for simultaneous nuclear segmentation and classification [46]. It is based on the encoded information within the horizontal and vertical map prediction of nuclear pixels, with respect to their center of mass. Then, the estimated distances were used to enhance the instance segmentation, especially for clustered nuclei. In [47], a self-supervised learning network, based on ResUnet-101, was proposed to reduce the requirement of manual annotations. The network achieved high accuracy but at the expense of computational complexity that also increases the execution time, as a result of applying Resnet101. All aforementioned instance segmentation models, except the last two, are designed based on binary classification. Hence, two different outputs should be predicted to distinguish nuclei boundary pixels. Also, most of them combine more than one network to enhance the segmentation accuracy. That, in turn, increases the computational complexity as well as the training/testing time. Besides, the training loss of some models that simultaneously predict more than one output is produced by accumulating the loss values, estimated for each different output. In most cases, only the overall training loss of such models is used for backpropagation (i.e., no backpropagation is done individually along each branch, for each different output, based on the specific training loss of that branch) that can mislead and complicate the training process and result in inaccurate model weights. Besides, only a handful number of the mentioned approaches can be considered generalized algorithms that use different types of tissues for training and testing.
In this paper, a real-time and accurate three-class nuclear segmentation deep learning algorithm is proposed. The three classes are the background, nuclei and boundary pixels. The algorithm is inspired by the Unet and residual learning nets. The main aim of the proposed algorithm is preserving the spatial features, while minimizing the processing time, in order to enhance the classification accuracy of the nuclear boundaries, generally, and the inter-nuclear boundaries, specifically. This is achieved by mildly downscaling the size of Feature Maps (FMs) while compacting their channels. Then, the compacted FMs are convolved by a proposed 2D convolution layer instead of the conventional 3D convolution layer. Consequently, the execution time is reduced. Also, as a result of the rational preservation of spatial features, a few number of layers are adequate to compensate the loss of spatial features. Hence, the computational complexity of the model and the execution time of both training and testing processes are dramatically reduced. The performance of the proposed model was evaluated by the AJI, F1-score and the FPS. A modification, as well, was made for the AJI to improve its accuracy of representing the actual object-level and pixel-level mismatch. The proposed algorithm was applied to multi-organ training and testing datasets, where some organs in the testing sets do not exist in the training set. The proposed algorithm achieved a high segmentation accuracy in real-time.
3 Proposed real-time nuclei segmentation model
The proposed architecture consists of three sequential steps: pre-processing, deep learning-based multiclass segmentation and post-processing, as described below.
3.1 Pre-processing
Since the H&E stained images of the training and testing datasets are produced by various labs and hospitals, then they are passed through different staining conditions. Consequently, color normalization should be applied to the input images as a preliminary step. All training and testing images are normalized based on the most widely used normalization method, presented by Macenko et al. [48]. To avoid overfitting, some augmentation techniques have been applied to the training set such as horizontal and vertical flipping, rotation with specific degrees, color jitter and random cropping with size of 256 × 256 that has the most important role and the greatest effect of preventing overfitting, when the size of feature maps is not downscaled intensively.
3.2 Real-time nuclei segmentation model
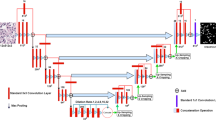
Inspired by Unet, a deep learning three-class segmentation model is proposed that classifies three pixel types: background, nuclei and boundary pixels to achieve the desired instance segmentation, using only one model and one training/testing step. The model consists of an encoder and a decoder, as shown in Fig. 1. In the conventional Unet architecture, the local and abstract features are extracted from high- and low- resolution maps, respectively, then they are combined together to retain fine and precise spatial features. Also, the number of channels of feature maps increases when getting deeper in the encoder to extract further important and complicated features, by combining formerly extracted features from previous layers. By increasing the number of feature maps, their 2-D dimensions (the length and width) are decreased for two reasons: first, to avoid the overfitting problem; and second, to minimize the computational complexity of the model that can cause the OOM problem for the utilized GPU. The OOM problem probably happens because the conventional 3D Convolution operation (Conv3) that is the core of any CNN-based architecture is carried out by applying all 3D-size kernel filters to the 3D-size feature maps, simultaneously (the third dimension of the kernel filter should match the number of channels of FMs), so that the element-wise matrix multiplication is concurrently performed for each channel of the input FMs. Afterwards, a horizontal aggregation of products of each individual element-wise matrix multiplication, and then a vertical aggregation along the depth dimension of the input FMs are carried out at each individual element in the FMs; to geberate new FMs (convolved FMs). The Conventional Size (Conv-Size) of data that are simultaneously stored in the RAM of GPU before aggregation, for each Conv3 layer, can be represented by (1); where B is the batch size, N is the number of channels of the input FMs (before convolution) that also represents the depth of each 3D kernel filter, M is the number of 3D kernel filters that also represents the number of convolved FMs (after convolution), hk and wk denote the height and width of the receptive field of each kernel filter, respectively, and h and w denote the height and width of the input FMs, respectively,
In conventional CNN-based architectures, generally, and U-net, specifically, the size of FMs is downscaled before convolution to reduce the computational complexity and the size of data, stored during convolution. However, the intensive size reduction of FMs can result in losing the tiny and fine spatial features, required to classify the minor-class pixels that are the nuclear boundary and inter-nuclear boundary pixels. This is why skip connections are added and a large number of layers are needed, in the conventional Unet, to partially offset the spatial feature loss. Nevertheless, an accurate instance segmentation, still, cannot be easily achieved based on the conventional architecture of Unet only, especially in the presence of overlapping and clustered nuclei. Moreover, increasing the number of layers, in turn, increases the processing time as well as the computational complexity.
In the proposed model, to reduce the computational complexity without degrading the segmentation accuracy, instead of convolving all channels of FMs by 3D kernel filters, they are first aggregated and compacted into one channel. This is done by the proposed Channels Sum (Ch-Sum) block, shown in Fig. 1. Then, the compacted channel is convolved by 2D kernel filters in the proposed 2D Convolutional layer (Conv2), as shown in Fig. 1. Compacting the input FMs and using 2D kernel filters produce the same result as the Conv3 layer. This is because the vertical aggregation and multiplication processes are just swapped, so that the pixels’ values are vertically aggregated at each position, along the depth dimension of FMs. Afterwards, the element-wise matrix multiplication and then the horizontal aggregation of element-wise matrix products are carried out. Consequently, the depth of the convolving kernel filters becomes one to match the depth of the compacted feature maps. The size of data that are simultaneously stored in the RAM of GPU for the proposed Conv2 process can be represented by (2). Comparing (2) with (1), it can be deduced that the number of multiplication operations is reduced by N for each individual Conv2 layer; that has a significant impact on the execution time:
In addition, compacting FMs enables increasing the number of kernel filters M, used in Conv2, to extract more features from the compacted FMs without increasing the computational complexity; that, in turn, enhances the segmentation accuracy. The proposed Ch_Sum operation is also applied on the inputs of skip connections, as shown in Fig. 1, to compact them before being concatenated and then convolved by the Conv2 layer in the up-sampling path (the decoder). Similarly, the proposed Ch-Sum and Conv2 blocks are used in the decoder, instead of Conv3, to minimize the execution time. Most state-of-the-art Unet-based architectures use residual learning nets as a backbone, especially Resnet50 that proved to extract more representative features [28]. In the proposed architecture, shown in Fig. 1, residual learning nets are also used. However, some modifications have been made, where the conventional Conv3 layer is replaced with the proposed Conv2 layer, to reduce the processing time and computational complexity. Figure 2 shows the structures of the modified residual blocks Res 1 and Res 2, developed for the proposed model, where chi-1 denotes the number of channels of input FMs before Conv2 layer, chi denotes the number of channels of extracted FMs after convolution and S denotes the stride value (used to downscale the size of FMs during convolution). In Fig. 2a, the input (identity map) of Res 1 is first merged into one channel, before the first Conv2 layer, while the 2D-size of that input is not dramatically reduced. Besides, the output of the Batch Normalization process (BN) in Res 1 is compressed into one channel. Then, it is added to the compacted identity map by the SUM operation (Fig. 2a), before being convolved by 2D kernel filters (instead of 3D filters), in the first Conv2 layer of Res 2 (Fig. 2b). Also, the output of the BN and ReLU processes in each of Res 1 and Res 2 blocks is first compressed into one channel, before being convolved by 2D kernel filters. The pseudocodes of the proposed Ch-Sum, Conv2, Res1 and Res2 architectures are stated in Algorithm1. It is worth noting that the BN, used in the proposed Conv2 layer is applied on the 3D-size FMs before being compacted by the Ch-Sum function for not degrading its efficiency. Based on several experiments, the number of layers, the size and the number of feature channels and the kernel size were empirically selected, according to the best performance of the validation set. Additionally, based on the time analysis, demonstrated in Sect. 5, the number of conventional Conv3 layers that can be replaced with the proposed Conv2 layers was determined. From Figs. 1 and 2, it can be deduced that the total number of convolution layers is 25 (including the transposed convolution), where 21 out of 25 convolution processes are implemented based on the proposed Conv2 layer; that notably reduces the computational complexity and the processing time of the proposed network, as demonstrated in Sect. 6.2. Also it can be noticed that the size of the input image is only downscaled twice, along the encoder path. Hence, only two transposed convolution processes (ConvT) are required in the decoder. Consequently, the accumulated error that results from using approximated up-sampling methods in ConvT layers is decreased. This can be considered another advantage of the moderate downscaling of the FMs size.
Another significant advantage is that the number of layers, added to compensate missing spatial features, is also reduced that in turn reduces the processing time; it can be noticed that only 14 layers, used in the encoder, are sufficient to provide better performance than some state-of-the-art architectures that use the standard Unet encoder together with Resnet50, as demonstrated in Sect. 6.2.
The applied loss function is defined by (3), where CCE is the Categorical Cross Entropy, IOUK is the Intersection Over Union [49] of the K-class pixels, and C is the number of classes (boundary, nucleus and background classes). The CCE loss function, defined by (4), is weighted by the class weight w(x) that is defined by (5) [27], to improve the instance segmentation result for boundary pixels (the minor class), where pl(x) is the Softmax output probability of the true class of Pixel x, NB is the total number of pixels in the batch size, wK(x), in (5), denotes the class weight of Pixel x to balance the class frequencies, d1 denotes the distance to the border of the nearest nucleus, and d2 is the distance to the border of the second nearest nucleus [27]. The class weight wK(x) was first initialized using (6), where NK is the total number of K-class pixels, and NKmin is the total number of pixels of the minor class. Then, the class-weights were gradually adapted, based on the validation loss results, up to 1, 1.5 and 5 for the background, nuclei and boundary pixels, respectively. The IOUK, defined by (7), is added to the loss function to penalize the predicted shapes of individual nuclei, where PK and MK are the matrices of the K-class predicted SoftMax probability and the K-class target mask, respectively:

Algorithm 1: Proposed 2D convolutional layer (Conv2) and RES1 and RES2 nets
3.3 Post-processing
It is observed that the nuclei boundaries, predicted by the proposed model, is thicker than the target nuclei boundaries. So, to enhance the segmentation quality of nuclei boundaries, some hypotheses have been set: only the outer pixels of the predicted boundary are considered as boundary pixels while the inner ones are considered as nuclei pixels. Additionally, any predicted nuclear segment, whose detected boundary is less than ½ the perimeter of that segment is considered as a background segment.
4 Datasets, evaluation metrics and implementation details
This section provides a detailed description of the applied datasets, the evaluation metrics, and the implementation details of the proposed model.
4.1 Datasets
For training, the multi-organ dataset, applied for training in the MoNuSeg 2018 challenge [23] is used. The dataset is originally released in [25] and it consists of 30 H&E stained tissue images, acquired from seven organs: colon, kidney, prostate, bladder, breast, liver and stomach. The images include malignant and benign cells, with manual annotations of the boundaries of 21,623 individual nuclei. The 30 images are downloaded from The Cancer Genome Atlas (TCGA) [50] that collected them from 18 hospitals. This guarantees appearance variation as a result of the difference in staining sources and scanners that can vary from lab to another. The dataset was then divided into 24 images and 6 images for training and validation, respectively. The size of the training images is 1000 × 1000 pixels, where each image was extracted from a separate WSI of a different patient, for diversity. For testing, two multi-organ datasets were used to evaluate the generalization capability of the proposed model. Both datasets are documented in the TCGA and acquired from different hospitals. They contain benign and malignant cells at different stages. The first dataset is the testing set, released in the MoNuSeg-2018 challenge [23]. It comprised 14 H&E images, taken from seven organs: bladder, brain, breast, colon, kidney, lung and prostate, where two organs: the lung and the brain are not included in the training and validation sets to assess the model generalization and robustness. The manual annotation of the boundaries of 7,223 individual nuclei is used in this work for model evaluation. The size of the testing images is 1000 × 1000 pixels, where each image was extracted from a separate WSI of a different patient. The second testing dataset combines the CPM-15 and CPM-17 datasets that are released in [51], and they comprised 25 and 32 H&E images, respectively. The images were taken from four organs: brain, neck, head and lungs that all are not included in the training and validation sets. The total number of manually annotated Nuclei is 2905 and 7570 for CPM-15 and CPM-17, respectively. The size of images varies from 400 × 400 to 1000 × 600 for CPM-15 dataset and from 500 × 500 to 600 × 600 for CPM-17 dataset. The combined dataset is known as Comb-CPM in the rest of the paper.
4.2 Evaluation metrics and the modified AJI
The instance segmentation quality of the model is evaluated based on two metrics: the first metric is the F1-score, defined by (8), where TP, FP and FN denote True Positive, False Positive and True Negative, respectively. The second metric is the Aggregated Jaccard Index (AJI), defined by (9) that is widely used to evaluate instance segmentation [25], where \({P}_{im}\) is the predicted nucleus that maximizes the pixel-wise Jaccard Index (JI), defined by (10), of the Ground Truth (GT) Nucleus \({G}_{i}\), and NGT is the number of ground truth nuclei. The main advantage of the AJI, introduced in [25], is that it is a parameter-free detection criterion that penalizes both the pixel- and object-level errors, regardless of the nuclear size and magnification. The authors, in [25], associated the same predicted nucleus to more than one GT nucleus as long as that predicted nucleus maximizes the JI of those GT nuclei. However, this violates logic, since each predicted nucleus should be associated to only one GT nucleus (if any). As a result, the numerator of the AJI formula, defined by (8), can falsely increase and would not represent the real quantitative value of intersection between the ground truth and predicted nuclei. Similarly, the first term of the denominator, in (8), can redundantly increase and would not accurately represent the quantitative value of union between the ground truth and predicted nuclei. Moreover, the second term of the denominator will not represent the real number of false negative cases (under-segmentation), if more than one GT nucleus is associated to the same predicted nucleus. To clarify that, Fig. 3 illustrates two sub-images, cropped from the test set, each one contains two GT nuclei (a and b in Fig. 3a, c, d in Fig. 3b) that are overlapping with the same predicted nucleus in each sub-figure, where the target and predicted boundaries of nuclei are represented with green and red-colored polygons, respectively. Logically, in Fig. 3a, the predicted nucleus should be associated to the GT Nucleus b, while the GT Nucleus a should be considered as a false negative case, since it just intersects with that predicted nucleus by an unnoticeable number of pixels. Similarly, in Fig. 3b, the predicted nucleus should be associated to the GT Nucleus d, while the GT Nucleus c should be considered as a false negative case. However, when the AJI algorithm is applied, it considers Nuclei a and c as true positive cases that violates logic. The problem of associating a predicted nucleus to multiple GT nuclei was addressed in [52], where the, Panoptic Quality (PQ) metric was proposed. A threshold is set for the JI, so that only the predicted nucleus that makes the JI of the ground truth nucleus ≥ 0.5 is selected. However, this returns the problem to be parameter dependent. Also, if the model fails to separate the closed or clustered nuclei (under-segmentation) the PQ metric my not detect that, especially in such cases where the size of adjacent/overlapping nuclei is appropriate as shown in Fig. 4, where the JI of the overlapping GT Nuclei a and b with the predicted nucleus are 0.512 and 0.601, respectively. Hence, according to the PQ, that predicted nucleus is falsely associated to both of them.
In this work, the original algorithm of the AJI has been modified to make it more logic and simulating to the human-way of thinking. The proposed Modified AJI (Mod-AJI), as well, addresses the mentioned problem of the PQ, so that if a detected nucleus is overlapping with more than one GT nucleus, it is associated to only one GT nucleus that maximizes the JI of that predicted nucleus. To achieve that, the modified algorithm of AJI loops over all detected nuclei. Then, not only the GT nuclei that do not intersect any predicted nucleus are considered false negative cases, but also any ground truth nucleus that intersects with one or more predicted nuclei, but it does not maximize the JI of any of them. The propose Mod-AJI is defined by (11). Applying the Mod-AJI on the examples, shown in Fig. 3, both GT Nuclei a and c are considered false negative, as should be. Also, by applying the Mod-AJI on the example, demonstrated in Fig. 4, the predicted nucleus is associated to GT Nucleus b, while GT Nucleus a is considered false negative:
where \({G}_{i}\) is the ground truth nucleus and \({P}_{j}\) is its associated segmented nucleus.
where \({G}_{im}\) is the ground truth nucleus that maximizes the pixel-wise JI of the predicted nucleus \({P}_{i}\), and \({M}_{GT}\) is the number of predicted nuclei. Although \(FN\) and \({FN}{\prime}\), presented in (9) and (11), respectively, denote the number of false negative cases, however they are not equal; \({FN}{\prime}\) represents the real false negative cases more precisely than \(FN\), as explained previously.
The proposed network was implemented on Nvidia Tesla T4 GPU. The batch size is 16, where 16 patches of size 256 × 256 were randomly cropped from each of training and validation sets. Then, the training patches were augmented to enhance the model robustness. The number of steps per epoch was 55, and Adam optimization was used with an initial learning rate of 102, and then it was reduced by a factor of 10 every time the validation loss stopped improving for 15 consecutive epochs.
4.3 Implementation details and training analysis
The early-stop strategy was employed to avoid overfitting. The training process took 2 h and 4 min, whereas the average testing time took 24 ms per 1000 × 1000 frame. In other words, the proposed model succeeded to segment nuclei in real-time as it can infer up to 41 FPS. Such a real-time performance is achieved as a result of replacing the 3D convolution layers with the proposed Conv2 layers, while using a few number of layers. Figure 5 illustrates the values of training and validation loss and accuracy of the implemented model, where the optimum values are 0 and 1 for the loss and accuracy, respectively. It can be noticed that the validation loss and accuracy fluctuate in the beginning, where the learning rate is relatively high. Also, it can be noticed that the values of validation loss and accuracy are slightly better than those of the training set, while approaching the point of stability. This is mainly because all augmentation techniques, mentioned in Sect. 3.1, were applied to the training set while only the random cropping was applied to the validation set.
5 Time analysis of the proposed and conventional convolution layers
A study was done to identify the impact of using the proposed compacted 2D convolution layers on the execution time. Both conventional 3D convolution and proposed convolution blocks, shown in Fig. 6, were implemented. Then, the execution time of each of them has been estimated, using Nvidia Titan 4 GPU, for different 2-D sizes (w and h) and number of channels (N) of the input FMs, and for different number of kernel filters (M). The receptive field of the kernel filters and the batch size are set to 3 × 3 and 16, respectively. Figure 6 illustrates the ratio between the estimated execution time of the compacted 2D convolution (TComp) and that of the conventional 3D convolution (TConv); that ratio reveals the effect of compacting feature maps, before convolution, on the execution time. It can be noticed from Fig. 7a that for 256 × 256 size input feature maps, M is not set to 1024 since it causes the OOM problem for the used GPU, even for the Conv2 layer. From Fig. 7, it can be deduced that the ratio TComp/TConv decreases by increasing the number of input feature maps (N) and their 2D dimensions (h and w). Also, the ratio indirectly decreases by increasing M, since the execution time of the element-wise matrix multiplication between the 3-D kernel filters and the input channels (N) in the conventional 3D convolution process increases by increasing the number of applied kernel filters (M), while it remains constant in case of the proposed compacted 2D convolution and becomes smaller than that of the conventional 3D convolution. This is because the 3-D kernel filters are replaced with 2-D kernel filters in the proposed Conv2 layer, as mentioned before, and they are applied only once on one compacted feature channel, instead of N feature channels. To recap, the effect of the compacted 2D convolution mainly depends on the number of input channels (N), output channels (M), the 2-D size of the input FMs, as well as the batch size and maybe the receptive field of the kernel filters. Hence, it is important to evaluate the efficiency of the proposed Conv2 layer before using it, based on these parameters. As aforementioned, in the proposed model the number of Conv2 layers are determined based on the time analysis, illustrated in Fig. 7.
6 Results
In this section, the qualitative and quantitative results of the proposed model, and the comparative results with some state-of-the-art models are presented and discussed.
6.1 Qualitative and quantitative analysis
The qualitative analysis of the proposed model is illustrated in Fig. 8 for seven sub-images, cropped from 7 different MoNuSeg-2018 testing images of 7 different organs, where two organs (the lungs and the brain) are not included in the training and validation sets, and the variation in the nuclear appearances and the H&E stain can be observed. Similarly, Fig. 9 illustrates the visual results of some sub-images, cropped from the Comb-CPM dataset. The nuclear boundary annotations and the corresponding segmented nuclei by the proposed model are listed in rows, in the figures. From Figs. 8 and 9, it can be noticed that the proposed model can successfully segment individual nuclei, regardless of the overlapping of some nuclei, and the diversity of tissue types, nuclear appearances and the H&E stain. It can be noticed also that although there is a number of false positive cases (over-segmentation) and false negative cases (under-segmentation), their number is unnoticeable, with respect to the total number of true positive cases.
For quantitative analysis, Table 1 lists the average values of AJI, Mod-AJI, F1-score and Mod-F1 score of the two testing datasets. The F1-score and Mod-F1 score are calculated individually based on the corresponding AJI and Mod-AJI, respectively, whose true positive, false positive and false negative cases are differently calculated, as explained before. Since the AJI/Mod-AJI penalizes both the pixel-level and object-level errors, while the F1-score/Mod-F1-score only penalizes the object-level error, the value of the later is always higher than that of the former. From Sect. 4.1, it can be deduced that the number of images of Comb-CPM testing set is quite larger than that of MoNuSeg-2018 testing set, and all its images belong to nuclei of completely unseen organs for the training and validation sets. Nevertheless, from Table 1, it can be noticed that the results of Comb-CPM testing set are comparable to those of MoNuSeg-2018 testing set; this indicates the generalization capability of the proposed model.
To analyze the difference between segmenting nuclei from seen and unseen organs more accurately, the AJI, Mod-AJI, F1-score and Mod-F1 score values of the 14 MoNuSeg-2018 testing images were calculated and listed in Table 2. Each image is identified by its organ type and sorted based on their AJI. Also, the average AJI, average Mod-AJI, average F1-score and average Mod-F1 score per organ are listed in Table 3. The organs denoted by the italics font in Tables 2 and 3 refer to the unseen organs that are not included in the training and validation sets. From Table 2, it can be observed that the AJI and Mod-AJI of all images are appropriate, except those of Image 3 (a lung image); this indicates that there are some ignored FN cases by the AJI that falsely affect both the numerator and denominator terms of the AJI, as explained before. Inspecting the average AJI and Mod-AJI, listed in Table 3, one can deduce that the lung and brain do not come in last place, although, they are not included in the training and validation sets. However, the order of the lung is not the same for the average AJI and Mod-AJI as a result of the incorrect FN cases, estimated by the AJI, for Image 3. From Tables 2 and 3, it can be noticed that the values of F1-score and Mod-F1-score of all images and organs are high, and approach the ideal value (that is one), even for the lung, whose F1-score came in last place. This indicates that the model is capable of separating overlapping nuclei, regardless of their appearance.
6.2 Comparison with state-of-the-art approaches
The proposed model was compared with some state-of-the-art architectures in terms of the segmentation accuracy, computational complexity and processing rate. To evaluate the segmentation accuracy, the overall AJI and F1-score of the proposed model and a number of alternative state-of-the-art architectures were obtained and listed in Table 4. The alternative models were trained and tested by MoNuSeg-2018 training and testing datasets, respectively, as the proposed model, for fair comparison. From Table 4, it can be noticed that the proposed model outperforms the first and second approaches, listed in Table 4, that use Resnet50 in their encoder. Also, its performance is comparable to the third approach that uses Resnet101 in its encoder, whereas the proposed model uses only 14 layers in the encoder path. This is because the proposed algorithm does not intensively reduce the size of FMs when getting deeper into the encoder, which moderately preserves the spatial features. Consequently, unlike the other methods, no large number of layers is required to compensate the deficiency of spatial features. Also, the moderate-scale reduction of the 2D size of FMs in the encoder of the proposed model reduced the required number of transposed convolution layers in the decoder, which in turn reduces the accumulated error that results from the up-sampling process, performed in such transposed convolution blocks. Regarding the computational complexity and the segmentation rate, the proposed model has been compared to other state-of-the-art architectures that are listed in Table 5, rather than those, listed in Table 4, as there is no sufficient information about either their inferring rate or the GPU, they used for implementation. For fair comparison, the proposed system was trained and tested on the training and testing datasets, released in [25], that were also used in the alternative approaches, listed in Table 5. The training set consists of 16 images, taken from 4 organs (liver, breast, prostate and kidney) of different patients and contain over 13,000 annotated nuclei. The testing set consists of 14 images of different patients that were taken from 7 organs, where three of them (stomach, bladder and colon) are not included in the training set. The third and fourth models, listed in Table 5, were re-implemented by Z. Cheng et al. [37], using the same training and testing sets, mentioned in this section, and the same GPU, used in [37]. For direct comparison with some alternative methods that are listed in Table 5, in terms of the segmentation quality, the Average Intersection over Union at threshold α (AIoU@ α), defined by (12) [53], was calculated beside the AJI and F1-score. The IoUi is defined by (13), where Gim is the GT nucleus that maximizes intersection over union of the predicted nucleus Pi. \(N_{\alpha }\) denotes the total number of predicted nuclei that satisfies IoUi ≥ α (α = 0.5 and 0.7). Based on the architectures, introduced in [34, 37, 38, 45] the computational complexity in terms of the number of convolution layers (that represents the number of layers of the neural network) has been approximately counted and listed in Table 5; this is to achieve a fair comparison, since the architectures were implemented on different GPUs. From Table 5 it can be noticed that the proposed architecture consists of the lowest number of convolution layers while its segmentation quality outperforms the first approach and is comparable to the other alternative methods, whose architectures are more complicated than itself.
Utilizing a small number of layers in the proposed model is a result of the rational preservation of spatial features, as mentioned before. On the contrary, the alternative architectures, listed in Table 5, combined multiple networks to compensate the loss in spatial features that is caused by the intensive downscaling of FMs. For instance, in [45], the deep residual inception network is used in the down sampling path while the decoder path of the standard Unet and the channel attention mechanisms were used for boundary and nuclei segmentation,
respectively. In [37], the authors combined the feature pyramid network and Resnet50 for ROI detection. In [38], the SSD was used to improve the ROI detection, and the ResNet50 was used as a backbone to enhance the segmentation accuracy. Also, the fully connected layers were used in the context refinement module that add more weights than Convolutional layers. Similarly, in [34], the SSD detector, the standard Unet and ResNet50 were used, while attention mechanisms were implemented to improve the detection and segmentation processes. In addition to the number of layers, the size of convolution layers also affects the computational complexity; unlike the alternative methods that only use the conventional 3D convolution layers, 21 out of 25 convolution layers in the proposed model are implemented, based on the proposed 2D convolutional layer, where the feature maps are compacted before convolution based on the study and time analysis, explained in Sect. 5. On the other hand, from Table 5 it can be noticed that the performance of utilized GPUs in the alternative models, in terms of the clock rate and Teraflop rate (TFLOPS), is better than that of Tesla T4 GPU, used in the proposed model. Nevertheless, the FPS of the proposed model that represents the segmentation rate is dramatically higher than those of the alternative approaches as a result of the reduction of computational complexity of the proposed model:
7 Conclusion
In this paper, a real-time and accurate deep learning algorithm for nuclear instance segmentation has been proposed. The proposed architecture is inspired by the Unet and residual nets. The main contribution of the proposed algorithm is replacing the conventional 3D convolutional layers, used in all CNN-based architectures, generally, and the Unet, specifically, with a proposed compacted 2D convolution layers. This is to fairly maintain the spatial features, without increasing the computational complexity, while increasing the segmentation rate and enhancing the instance segmentation of boundary pixels. This is achieved by a reasonable downscaling of the size of feature maps and compacting their channels, then they are convolved by 2D kernel filters, instead of the conventional 3D kernel filters, inside and outside the residual learning nets. As a result, the segmentation speed is dramatically increased and the total number of layers, required to compensate the loss of spatial features, is reduced that in turn minimizes the computational complexity and the overall processing time. Moreover, the accumulated error that results from up-sampling operations in the decoder is reduced as a result of the moderate reduction of the size of feature maps in the encoder. Consequently, the training time, required to achieve the optimum weights of the model is decreased. Also, the AJI formula has been modified to enhance its accuracy of representing the object- level and pixel- level errors. The proposed algorithm succeeded to segment nuclei of organs that are not included in the training dataset and its performance outperformed some alternative state-of-the-art approaches, whose architectures are more complicated than that of the proposed model. Moreover, not only the FPS of the proposed model is considered the highest among some state-of-the-art approaches that are concerned with increasing the processing rate as well as the segmentation accuracy, but it also achieved a real-time segmentation rate. In general, the proposed compacted 2D convolution layer can replace the conventional 3D convolution layer in any CNN-based application that is concerned with increasing the processing rate and minimizing the computational complexity.
In the future, the proposed model would be applied on further datasets that contain images of multiple microscopic resolution and different types of stains that would require some modifications on the proposed algorithm to enhance its robustness and make it more generalized.
Data availability
The code developed for the proposed deep learning model in this paper is now being used in another research project. After completion of that project, it will be publicly accessible.
References
Irshad, H., Veillard, A., Roux, L., Racoceanu, D.: Methods for nuclei detection, segmentation, and classification in digital histopathology: a review. IEEE Rev. Biomed. Eng. 7, 97–114 (2014)
Rittscher, J.: Characterization of biological processes through automated image analysis. J. Annu. Rev. Biomed. Eng. 12(1), 315–344 (2010)
Xue, Y., Ray, N., Hugh, J., Bigras, G.: A novel framework to integrate convolutional neural network with compressed sensing for cell detection. In: 2017 IEEE Int. Conf. on Image Processing (ICIP)., pp. 2319–2323( 2017)
Naik, S., Doyle, S., Agner, S., Madabhushi, A., et al.: Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In: 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro., pp. 284–287 (2008)
Gurcan, M.N., et al.: Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2, 147–171 (2009)
Llewellyn, H.: Observer variation, dysplasia grading, and hpv typing: a review. Am. J. Clin. Pathol. 114, 21–35 (2000)
Louis, D.N., et al.: Computational pathology: a path ahead. Arch. Pathol. Lab. Med. 140(1), 41–50 (2015)
Filipczuk, P., Fevens, et al.: Computer-aided breast cancer diagnosis based on the analysis of cytological images of fine needle biopsies. IEEE Trans. Med. Imaging 32(12), 2169–2178 (2013)
Sethi, A., Sha, L.: Computational pathology for predicting prostate cancer recurrence. In: Proceedings of AACR 106th annual meeting. (2015)
Xue, J.H., Titterington, D.M.: t-tests, F-tests and Otsu’s methods for image thresholding. IEEE Trans. Image Process. 20(8), 2392–2396 (2011)
Yang, X., et al.: Nuclei segmentation using marker controlled watershed, tracking using mean-shift, and Kalman filter in time-lapse microscopy. IEEE Trans. Circuits Syst. 53(11), 2405–2414 (2006)
Al-Kofahi, Y., et al.: Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans. Biomed. Eng. 57(4), 841–852 (2010)
Ali, S., Madabhushi, A.: An integrated region-, boundary-, shape based active contour for multiple object overlap resolution in histological imagery. IEEE Trans. Med. Imag. 31(7), 1448–1460 (2012)
Wienert, S., et al.: Detection and segmentation of cell nuclei in virtual microscopy images: a minimum-model approach. Sci. Rep. 503, 1–7 (2012). https://doi.org/10.1038/srep00503
Moen, E., et al.: Deep learning for cellular image analysis. J. Nat. Methods 16(12), 1233–1246 (2019)
Fatichah, C., Tangel, M.L., Rahmat, M.: Optimization of local fuzzy patterns based on fuzzy contrast measure for white blood cell texture feature extraction. J. Adv. Comput. Intell. Intell. Inform. 16(3), 412–419 (2012)
Chen, S., et al.: Recent advances in morphological cell image analysis. Comput. Math. Methods Med. 2012(24), 1–10 (2012). https://doi.org/10.1155/2012/101536
Mattie, M., et al.: Content based cell image retrieval using automated feature extraction. J. Am. Med. Inform. Assoc. 7(4), 404–415 (2000)
Mao, K.Z., Zhao, P., Tan, P.: Supervised learning based cell image segmentation for p53 immunohistochemistry. IEEE Trans. Biomed. Eng. 53(6), 1153–1163 (2006)
Paul, P., Bhowmik, M., Bhattacharjee, D.: Automated cervical cancer detection using pap smear images. In: Springer 4th Int. Conf. on Soft Computing for Problem Solving., pp. 267–278 (2015)
Bora, K., Chowdhury, M., Mahanta, L., Kundu, M., Das, A.: Automated classification of pap smear images to detect cervical dysplasia. Comput. Methods Programs Biomed. 138, 31–47 (2017)
Liu, Z., et al.: A survey on applications of deep learning in microscopy image analysis. Comput. Biol. Med. 134, 104523–104535 (2021). https://doi.org/10.1016/j.compbiomed.2021.104523
Kumar, N., et al.: A multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imag. 39, 1380–1391 (2019)
Verma, R., Kumar, et al.: MoNuSAC2020: a multi-organ nuclei segmentation and classification challenge. IEEE Trans. Med. Imaging 41(4), 3413–3423 (2021)
Kumar, N., Verma, R., Sharma, S., Bhargava, S., et al.: A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 36, 1550–1560 (2017)
Zhao, B., et al.: Triple Unet: hematoxylin-aware nuclei segmentation with progressive dense feature aggregation. Med. Image Anal. 65(22), 101786 (2020)
Ronneberger, O., Fischer, P., Brox, T.: Unet: Convolutional networks for biomedical image segmentation. In: MICCAI. Springe.
He, K., et al.: Deep residual learning for image recognition. In: 2016 IEEE Conf. on Computer Vision and Pattern Recognition., pp. 770–77 8(2016)
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., et al.: Single shot multibox detector. In: European Conference on Computer Vision., pp. 21–37 (2016)
Girshick, R.: Fast R-CNN. 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1440–1448 (2015)
Ren, S., He, K., Girshick, R., Sun, J.: Faster r-cnn: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39(6), 1137–1149 (2017)
He, K., Gkioxari, G., Dollar, P., Girshick, R.: Mask r-cnn. In: 2017 IEEE Int. Conf. on Computer Vision (ICCV), 2980–2988 (2017)
Lin, T., et al.: Focal loss for dense object detection. In: Proceedings of the IEEE International Conf. on Computer Vision. Pp. 2980–2988 (2017)
Yi, J., et al.: Attentive neural cell instance segmentation. Med. Image Anal. 55, 228–240 (2019)
Johnson, J.: Adapting mask-rcnn for automatic nucleus segmentation. In: Proceedings of the Computer Vision Conference. (2019)
Liang, H., et al.: A region-based convolutional network for nuclei detection and segmentation in microscopy images. Biomed. Signal Process. Control, 71, 1–14 (2022). https://doi.org/10.1016/j.bspc.2021.1032
Cheng, Z., Qu, A.: Fast and Accurate Algorithm for Nuclei Instance Segmentation in Microscopy Images. IEEE Access 8, 158679–158689 (2020)
Yi, J., Wu, P., Huang, Q., et al.: Context-refined neural cell instance segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 1028–1032 (2019)
Wang, R., Kamata, S.: Stain-Refinement and Boundary-Enhancement Weight Maps for Multi-organ Nuclei Segmentation. In: 4th International Conference on Imaging, Vision & Pattern Recognition, pp. 1–7 (2021)
Rahmon, G., et al.: Extending Unet Network for Improved Nuclei Instance Segmentation Accuracy in Histopathology Images. IEEE Applied Imagery Pattern Recognition Workshop. 1–7(2021)
Zhou, Z., et al.: Synthesis and segmentation method of cross- staining style nuclei pathology image based on adversarial learning. In: IEEE Intl Conf. on Parallel & Distributed Processing with Applications. (2021)
Hu, W., Sheng, H., Wu, J., et al.: Generative adversarial training for weakly supervised nuclei instance segmentation. In: 2020 IEEE Int. Conf. on Systems Man and Cybernetics (SMC), pp. 3649–3654 (2020)
Zhou, C., et al.: Recursive training strategy for a deep learning network for segmentation of pathology nuclei with incomplete annotation. IEEE Access. 10, 49337–49346 (2022)
Mahbod, A., et al.: CryoNuSeg: a dataset for nuclei instance segmentation of cryosectioned H&E-stained histological images. Comput. Biol. Med. 132, 104349 (2021)
Zeng, Z., Xie, W., Zhang, Y., Lu, Y.: RIC-Unet: an improved neural network based on Unet for nuclei segmentation in histology images. IEEE Access Comput. Sci. 7, 21420–21428 (2019)
Graham, S., et al.: HoVer-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563–101580 (2019)
Xie, X., et al.: Instance-aware self-supervised learning for nuclei segmentation. In: Martel, A.L., et al. (ed.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol. 12256, pp. 341–350. Springer, Cham (2020).
Macenko, M., et al.: A method for normalizing histology slides for quantitative analysis. Proceedings of the IEEE Int. Symp. Biomedical Imaging: From Nano to Macro, 1107–1110 (2009)
Jaccard, P.: Etude comparative de la distribution florale dans une portiondes alpes et des jura. Bull Soc Vaudoise Sci Nat. 37(142), 547–579 (1901)
The cancer genome atlas (tcga). http://cancergenome.nih.gov/. Accessed 20 Jan 2024
Quoc, V., et al.: Methods for segmentation and classification of digital microscopy tissue images. Front. Bioeng. Biotechnol. 7, 1–15 (2019). https://doi.org/10.3389/fbioe.2019.00053
Kirillov, A., et al.: Panoptic segmentation. CoRR, vol. abs/1801.00868, 2018. [Online]. Available: http://arxiv.org/abs/1801.00868
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., et. al.: Ssd: Single shot multibox detector. In ECCV. Springer, pp. 21–37 (2016)
Acknowledgements
The author would like to thank Dr. Neamat Hanafi, a professor of cell biology and histology at the EAEA, for giving a hand to understand the histology images of nuclei and realize the pathologists’ way of thinking to analyze and segment such kind of images that helped the author while implementing the model and modifying the AJI.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
As it is a single- author paper, all contributions have been done by Noha. Y. Ahmed.
Corresponding author
Ethics declarations
Conflict of interest
The author states that there is no conflict of interest and there is no financial nor personal relationship with any individuals or organizations that can inappropriately influence the proposed research work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahmed, N.Y. Real-time and accurate deep learning-based multi-organ nucleus segmentation in histology images. J Real-Time Image Proc 21, 43 (2024). https://doi.org/10.1007/s11554-024-01420-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11554-024-01420-0