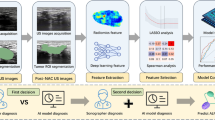
Abstract
Objectives
The study aimed to develop a combined model that integrates deep learning (DL), radiomics, and clinical data to classify lung nodules into benign or malignant categories, and to further classify lung nodules into different pathological subtypes and Lung Imaging Reporting and Data System (Lung-RADS) scores.
Materials and methods
The proposed model was trained, validated, and tested using three datasets: one public dataset, the Lung Nodule Analysis 2016 (LUNA16) Grand challenge dataset (n = 1004), and two private datasets, the Lung Nodule Received Operation (LNOP) dataset (n = 1027) and the Lung Nodule in Health Examination (LNHE) dataset (n = 1525). The proposed model used a stacked ensemble model by employing a machine learning (ML) approach with an AutoGluon-Tabular classifier. The input variables were modified 3D convolutional neural network (CNN) features, radiomics features, and clinical features. Three classification tasks were performed: Task 1: Classification of lung nodules into benign or malignant in the LUNA16 dataset; Task 2: Classification of lung nodules into different pathological subtypes; and Task 3: Classification of Lung-RADS score. Classification performance was determined based on accuracy, recall, precision, and F1-score. Ten-fold cross-validation was applied to each task.
Results
The proposed model achieved high accuracy in classifying lung nodules into benign or malignant categories in LUNA 16 with an accuracy of 92.8%, as well as in classifying lung nodules into different pathological subtypes with an F1-score of 75.5% and Lung-RADS scores with an F1-score of 80.4%.
Conclusion
Our proposed model provides an accurate classification of lung nodules based on the benign/malignant, different pathological subtypes, and Lung-RADS system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-contrast low-dose chest computed tomography (LDCT) is the standard imaging modality for lung cancer screening [1]. Based on the National Lung Screening Trial (NLST), screening LDCT can reduce mortality by 20% in the high-risk group compared with screening chest radiography [2, 3]. The use of screening chest CT is increasing, but false-positive and overdiagnosis rate are not negligible [4, 5]. In the NLST, only 3.8% of positive results were diagnosed as lung cancer [3]. In a previous study conducted at a tertiary referral center in Taiwan, we showed that 45% of resected small lung nodules of < 6 mm were benign [6].
The management of indeterminate pulmonary nodules (IPNs) is difficult [7] because most IPNs are benign [8]. Clinicians must accurately assess the risk of malignancy in order to diagnose and treat cancerous lesions without performing unnecessary tests and procedures in patients with benign nodules in a timely manner [9]. Lung-RADS, introduced by the American College of Radiology (ACR), categorizes nodules into five groups based on their risk of malignancy. Categories 1 (negative) and 2 (benign appearance) are considered negative and undergo annual screening. Categories 3 (probably benign) and 4A/4B/4X (suspicious) are considered positive and require additional evaluation before the next annual screening. Lung-RADS uses a 6 mm threshold, which reduces false positives without delaying lung cancer diagnosis compared to the 4 mm threshold used in the NLST [10, 11]. Although guidelines for nodule management are available, accurate characterization of IPNs remains tedious and subject to inter- and intra-reader variability.
Adenocarcinoma is the most common histologic subtype of lung cancer [12]. Atypical adenomatous hyperplasia (AAH) may be a precursor lesion of adenocarcinoma [13]. The invasiveness of lung adenocarcinoma is assessed by a multidisciplinary classification [10], categorizing it as adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), or invasive adenocarcinoma (IA). Given the central role of diagnosis in treatment and prognosis, invasiveness has a significant impact on survival [14, 15]. Improving the prediction of invasiveness by chest CT offers significant clinical benefit to patients with lung cancer.
Computer-aided detection (CAD) in chest CT has long been recognized for its ability to improve sensitivity in nodule detection [16, 17]. Recent breakthroughs in deep learning (DL) for medical imaging have expanded its capabilities to include automatic nodule segmentation [18], classification [19], nodule measurement, and malignancy risk assessment [20]. However, most of the previous studies were conducted under conditions that differ from real-world practice and often selected for disease prevalence and dichotomized distribution. In addition, current CAD cannot predict lung nodule pathology preoperatively. Therefore, there is a significant need for studies that evaluate artificial intelligence (AI) models in real-world populations and provide preoperative prediction of lung nodule pathology to guide clinical decision making.
Lung nodules can be classified using two basic approaches: (1) the radiomic feature extraction from chest CT scans, either 2D or 3D [21, 22], and (2) convolutional neural networks (CNN) [23, 24]. Many recent studies have used these tools to predict the invasiveness of lung nodules [23, 25,26,27,28,29,30]. The radiomics approach requires an appropriate lung segmentation and feature extraction algorithm to classify the tumor, while CNN does not need such an algorithm, but requires a huge dataset [20]. In this study, we aimed to investigate the diagnostic performance of our proposed AI model in open dataset, and private dataset (both surgery and health checkup participants). The purpose of our study was to investigate whether our proposed combined AI model (integrating DL, radiomics, and clinical data) could improve the classification of lung nodules into benign/malignant, histological types and Lung-RADS categorization.
Materials and methods
This retrospective study was approved by the institutional review board of National Cheng Kung University Hospital (A-ER-108–359) and the requirement for written informed consent was waived because the data were analyzed retrospectively and anonymously.
Datasets
The following three datasets were used:
-
(1)
Lung Nodule Analysis 2016 (LUNA16) Grand Challenge dataset [31]: This is a publicly available dataset containing1186 lung nodules from 888 patients.
-
(2)
Lung Nodule Received Operation (LNOP) dataset: It includes 1027 lung nodules from 708 patients who underwent surgical resection with histopathological diagnosis at the National Cheng Kung University Hospital (NCKUH) between December 2018 and December 2021.
-
(3)
Lung Nodule in Health Examination (LNHE) dataset: It includes 1525 lung nodules from 653 patients, which were found during a healthy examination between January 2019 and December 2021 at the NCKUH.
Figure 1 shows how the datasets were partitioned for each task. Ten-fold cross-validation was applied to each task. The LNOP dataset also includes clinical information that have been observed to be associated with lung cancer and tumor phenotype [32,33,34,35,36], such as age, sex, smoking history (defined as positive smoking history regardless of whether the patient is an active smoker or has quit smoking) and the presence of a family history of lung cancer in first-degree relatives.
Classification tasks
We performed three classification tasks in this study:
-
(1)
Task 1: Classification of lung nodules as benign or malignant in the LUNA16 dataset.
-
(2)
Task 2.1: Three-class classification of
-
(i)
IA
-
(ii)
MIA + AIS
-
(iii)
AAH + other benign lesions.
Task 2.2: Four class classification of AAH, AIS, MIA, an d IA.
-
(i)
-
(3)
Task 3: Four-class classification of Lung-RADS score: 2, 3, 4A, 4B + 4X.
Task 2.1 was designed to resemble the real clinical scenario based on treatment strategies: (i) IA has the worst prognosis, and lobectomy is often recommended [37], (ii) MIA + AIS have almost 100% survival probability, and limited resections are suggested [37, 38], (iii) AAH + other benign lesions required conservative treatment or follow-up. Task 2.2 provided four-class classification of adenocarcinoma spectrum lesions from pre-invasive to invasive lesions into AAH, AIS, MIA, and IA. Only the LNOP dataset was used in Task 2, whereas the LNOP and LNHE datasets were used in Task 3. Squamous cell carcinoma (SqCC) and metastasis were excluded in Task 2.1 and Task 2.2 due to their rarity in the lung screening program. To balance the data, only partial data were used in Tasks 2.2 and 3.
Image acquisition
In the LUNA16 dataset, the size of all the nodules was greater than 3 mm and slice thickness was less than 2.5 mm. In the LNOP and LNHE datasets, all CT images were acquired using the Siemens SOMATOM Definition Flash, Siemens SOMATOM Definition AS, SOMATOM Definition Edge, and GE Optima CT660. The CT protocols were as follows: 120 kVp; tube current, 150–200 mA with automatic tube current modulation in the LNOP dataset and 30 mA in the LNHE dataset. The slice thickness ranged from 0.625 to 1.5 mm, and the image size was 512 × 512 pixels.
Image preprocessing
The image preprocessing module included (1) voxel resampling, (2) cropping, and (3) Hounsfield unit conversion. Because the voxel spacing of each CT image might be different, we resampled the voxel spacing of all CT images and masks to the smallest spacing value in the dataset (0.48 × 0.48 × 0.625 mm3). The 3D CT images were then cropped to the size of 32 × 32 × 32 as the input to the DL model. Finally, the Hounsfield unit in the range between -1024 and 400 was converted to a decimal between 0 and 1 and stored in the single precision floating point format. This is a normalization of the input data for the neural network.
Radiomic feature extraction
Contours defining the 3D tumor region of interest were manually drawn slice by slice on axial images after a consensus was reached between the thoracic radiologist (C.Y.L., with 10 years of experience) and the thoracic surgeon (C.C.C., with 10 years of experience). The thoracic radiologist blinded to the clinicopathologic data performed tumor segmentation by using graphical user interface (GUI) written in Python. Tumor delineation was performed in a lung window setting to highlight lung structures on the axial CT plane, including bronchi, blood vessels, and vacuoles within the nodules and excluding irrelevant normal lung tissue, mediastinal structures, and chest wall, as shown in Fig. 2.
Graphical user interface for the lung nodule segmentation. A Lung nodule segmentation is manually performed by a thoracic radiologist. B A bounding box created from a segmentation mask automatically. C A segmentation mask is provided automatically. D The original non-enhanced axial CT images in lung window setting shows an irregular left upper lobe (LUL) mass
A total of 1319 radiomic features were extracted by using Pyradiomics [39] with two image filters, the Laplacian of Gaussian (LoG) filter and the wavelet filter to highlight specific features [41], and six feature families, including shape features, first-order statistics, and texture features (gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), and gray level dependence matrix (GLDM)) [40,41,42].
DL model
The DL models were derived from a modified 3D CNN, NASLung [43]. To improve the model performance, we made two modifications. First, we replaced the convolutional block attention module (CBAM) with a coordinate attention (CA) block to ensure that the model efficiently captures long-range features with accurate location information [44, 45]. Second, the residual block in NASLung was adapted to be applicable to lung nodule classification, inspiration by the dilated residual dense block (DRDB) [46]. The training environment and strategy were detailed in Appendix S1.
Development and combination of models for lesion classification
The global framework is shown in Fig. 3. To combine the DL and radiomics models, we applied a stacked ensemble model using a machine learning (ML) approach with the AutoGluon-Tabular classifier [42]. Autogluon is an open-source automated ML library developed by the Amazon Web Services (AWS). It serves as a framework for automating ML tasks, allowing users to automatically select and train ML models. In AutoGluon-Tabular, it integrates several basic ML models, including neural networks, LightGBM boosted trees, CatBoost boosted trees, Random Forests, Extremely Randomized Trees, and K-Nearest Neighbors algorithm. For each ML model selected, Autogluon optimizes the training process by automating hyperparameter tuning, achieving superior performance by eliminating manual iteration through hyperparameter configurations. Additionally, it uses multi-layer stacking strategies to enhance prediction performance. The ensemble model provided a probability of malignancy (Task 1), a histopathology result (Task 2.1 and 2.2), and a Lung-RADS score (Task 3). To test whether the clinical features had any additional predictive value for histopathologic subtype classification, the ensemble model was retrained after the addition of clinical features for Task 2.1 and 2.2.
Data processing pipeline. The parallel radiomics and DL model encodes the input images to features which be combine with clinical parameters. Then the combined features be classified by an ensemble classification model. The methods denoted as L, M, and N in modified NASLung architecture were determined based on the original NASLung framework
Radiologist reading
Target lesions were independently identified and classified according to Lung-RADS version 1.1 by a thoracic radiologist (C.Y.L., with 10 years of experience) who was blinded to all patient clinical and demographic information. In addition, the imaging and histopathologic results of each patient in the LNOP dataset were reviewed by a multidisciplinary thoracic tumor board as a standard of care.
Statistics
Continuous variables were reported as the mean ± standard deviation, and categorical variables were reported as n (%). In the ten-fold cross-validation, the results were presented as mean accuracies. To verify the performance of the different classification models, we calculated the accuracy, macro-recall, macro-precision, and macro-F1 scores. Accuracy is the ratio of the correctly classified samples to the total samples. The precision is the ratio of the true positive results to all positive results, and the average of the precision of each sample label is the macro-precision. The recall is defined as true-positive results divided by the sum of true-positive and false-negative results, and the mean value of the recall of each sample label is the macro-recall. The macro-F1 score is the harmonic mean of the macro-precision and macro-recall.
Results
Dataset characteristics
Figure 4 shows the 3D diameter distribution and percentage of solid components in lung nodules from the LUNA16, LNOP, and LNHE datasets. Clinical and pathologic characteristics are detailed in Table 1. Females comprised the majority of patients in both the LNOP (64.5%) and LNHE (53.4%) groups. The mean age of the patients was 59.6 years in the LNOP group and 54.7 years in the LNHE group. Among the patients in the LNOP group, 27.8% had a smoking history, while 16.0% had a family history of lung cancer in first-degree relatives. IA was the most common pathology among patients in the LNOP group, accounting for 25.1% of all malignancies. According to Lung-RADS, 36.5% of the patients in the LNOP group were classified as 4A + 4B + 4X. In contrast, because the patients in the LNHE group were all asymptomatic individuals undergoing health screening, only 9.1% were classified as 4A + 4B + 4X.
In terms of 3D diameter, the majority of nodules in the LUNA16 and LNHE datasets were smaller than 30 mm, while LNOP had a wider distribution with a maximum of 53 mm. The wider distribution of nodule sizes in the LNOP dataset may be attributed to the fact that nodules deemed suitable for surgical removal were more likely to be malignant based on the subjective judgment of the clinicians. Regarding the solid component, the LUNA16 dataset had a higher proportion of solid components compared to the LNOP and LNHE datasets. This discrepancy may be due to the fact that the LNOP and LNHE datasets consisted primarily of individuals of Asian descent, who had a higher prevalence of ground-glass opacities.
Classification of lesions as benign or malignant
Our classification model had an accuracy of 92.80% in ten-fold cross-validation and an F1-score of 92.16% (Table 2). The confusion matrix of the LUNA16 classification is shown in Fig. 5A. The accuracy of the benign lesion was 96.17% and the accuracy of the malignant lesion was 89.43%.
In the ablation study for Task 1, we evaluated the effectiveness of modified NASLung and the incorporation of radiomics features. In the DL model, we incrementally added image preprocessing, replaced the CBAM and residual block in the original NASLung with CA and DRDB, and used the AutoGluon-Tabular classifier. This ultimately increased the accuracy from 88.78% to 92.21% and increased the F1-score from 87.92% to 91.61%. In the radiomics model, after image preprocessing and applying the AutoGluon-Tabular classifier, using all radiomics features showed the best result with an accuracy of 90.75%, F1-score 89.86. However, the result was inferior to the DL model. The combination of DL model and radiomics model showed the best result, with an accuracy of 92.80%, F1-score 92.16% (Table 3).
Three-class classification of IA, MIA + AIS, AAH + other benign lesions
The F1-score and the overall accuracy of Task 2.1 were 75.45% and 74.76%, respectively (Table 2). The confusion matrix of Task 2.1 is shown in Fig. 5B. The accuracy of IA, AIS/MIA, and AAH/others was 84.70%, 74.45% and 67.82%, respectively.
In the ablation study for Task 2.1, we investigated the effect of clinical features on the prediction of malignancy in lung nodules. After incorporating four clinical features, namely smoking history, family history, age, and sex, we observed an increase in the accuracy from 72.87% to 74.76% and in the macro F1-score from 73.57% to 75.45% (Table S1).
Four-class classification of AAH, AIS, MIA, and IA
The F1-score and the overall accuracy of Task 2.2 were 68.70% and 68.52%, respectively (Table 2). The confusion matrix of Task 2.2 is shown in Fig. 5C. The accuracies of IA, MIA, AIS, and AAH were 87.33%, 61.88%, 57.06% and 68.27%, respectively. The model performed better in distinguishing AAH from IA, but showed poorer performance in distinguishing AIS from MIA.
Four-class classification of lung-RADS score 2, 3, 4A, 4B + 4X
The F1-score and the overall accuracy of Task 3 were 80.38% and 80.48%, respectively (Table 2). The confusion matrix of Task 3 is shown in Fig. 5D. The accuracies of Lung-RADS 4B + 4X, 4A, 3, and 2 were 93.34%, 77.56%, 64.76% and 86.19%, respectively. In addition, the relationship between final pathology and Lung-RADS score in the LNOP dataset was presented in Table S2. The LNOP dataset showed a higher risk of malignancy when using Lung-RADS categorization as a reference (Lung-RADS score 2: 39.8%, score 3: 70.9%, score 4A: 77.3%, score 4B + 4X: 80%).
Discussion
We proposed a combined model to classify lung nodules using radiomics and DL. In addition, we evaluated the added value of clinical features to classify different pathologic subtypes of lung nodule. Our fusion model achieved high predictive accuracy and outperformed all other single method-based models in the LUNA16 dataset after module modification. We achieved F1-scores of 75.45% and 68.7% in three- and four-class classifications, respectively, when predicting pathological subtypes using private datasets. Classification of the Lung-RADS score gave an accuracy of 80.38%. Our proposed model demonstrated high accuracy in classifying lung nodules as benign or malignant, as well as in classifying lung nodules into different pathological subtypes and Lung-RADS scores.
The result of the ablation study of Task 2.1 shows that clinical data play an important role in the classification of pathological subtypes of lung nodules, increasing the classification accuracy by 1.89%. Smoking has the greatest impact on classification, increasing the accuracy by 0.94% (Table S1). In our study, the LNOP dataset showed a higher risk of malignancy when the Lung-RADS categorization was used as a reference. This may be explained by differences in ethnicity, as Asian populations with adenocarcinoma often presented with ground-glass opacities. This highlights the inadequacy of relying on Lung-RADS alone for follow-up and management decisions. Our AI models could lead to more personalized treatment plans by better predicting the pathological nature of nodules. In cases with incorrect pathological subtype classification but correct Lung-RADS categorization, we found that the inconsistency of solid parts and pathological report is the cause of the suboptimal result of pathological subtype classification (Fig. 6). The spectrum of early-stage lung cancer shows progressive size and density changes on chest CT, making it difficult to accurately differentiate IA, MIA, AIS, and AAH. In Task 2.1, the classification of the AAH/other group has the lowest accuracy because this group also includes other benign lesions, such as tuberculosis (TB), cryptococcal infection, and organizing pneumonia. TB presents with various imaging features, including cavitation, pleural tethering, and spiculation, which can mimic malignancy. As a result, it not only poses a clinical diagnostic problem but also affects the learning effect of the model. Infection and inflammation may appear as ground-glass opacities in the early stages of CT imaging. This can only be confirmed after serial follow-ups and clinical correlations. However, even with these confounding cases in our dataset, the accuracy of predicting IA was 84.70%. After eliminating these confusing benign lesions, the accuracy of IA prediction can increase to 87.33% in Task 2.2 (Fig. 5).
Representations examples of pathology misclassification followed by accurate Lung-RADS categorization using a chest CT AI classification model. A Initial misclassification as MIA, later confirmed as AAH upon pathology examination. Correctly classified as Lung-RADS category 3. B Initial misclassification as AAH, later confirmed as AIS upon pathology examination. Correctly classified as Lung-RADS category 2. C Initial misclassification as IA, later confirmed as TB upon pathology examination. Correctly classified as Lung-RADS category 4B + 4X. D Initial misclassification as IA, later confirmed as organizing pneumonia upon pathology examination. Correctly classified as Lung-RADS category 4B + 4X
Recent medical literature has demonstrated an emerging trend in the use of DL models for the diagnosis of lung cancer. A literature review of previous studies is summarized in Table 4. Qi et al. achieved an accuracy of 76.9% and a F1 score of 60.9% in a dataset of 448 pure ground glass nodules (GGN), categorizing them as IA or other types [47]. Zhang et al. excelled with an 89.8% accuracy in distinguishing malignant from benign nodules in a dataset of 972 patients after excluding nodules that were difficult to segment [48]. Notably, GGNs are inherently challenging due to their blurred borders, potentially containing malignancies such as AIS. Liu et al. achieved an 81.6% accuracy in a binary malignant versus benign classification in 204 patients [49], while Marappan et al. achieved a 76.67% accuracy in distinguishing MIA from IA in a dataset of 105 patients [50]. Qi et al. extended their study to 417 patients and classified nodules as small cell lung cancer (SCLC), IA, and SqCC. They reported individual accuracies of 0.83, 0.75, and 0.67 for SCLC, IA, and SqCC, respectively, with an average accuracy of 0.75. The weighted F1-average was also 0.75 [51]. Notably, SCLC and SqCC were predominantly observed as pure solid tumors, whereas IA showed a broader distribution from pure GGN to pure solid patterns. Kao et al. focused on pure GGNs in a dataset of 338 patients and achieved an accuracy of 70.6% in differentiating AIS + MIA from IA [52]. These studies used private datasets with specific limitations, including nodule size, solid or ground-glass opacity components, which may impact the applicability of the models. It is well known that the performance of a model can vary across different datasets, making it difficult to generalize results from private datasets to broader clinical scenarios. In this context, we present a quantitative comparison between our private and publicly available datasets, focusing on nodule size and solid component distributions (Fig. 4). This analysis aims to provide a clearer understanding of the differences between our private dataset and public datasets, allowing for an informed assessment of the suitability of the model for clinical practice or research in specific medical domains.
CAD has been used for Lung-RADS categorization. Park et al. used CAD to improve the inter-reader agreement of Lung-RADS category, from moderate to substantial with CAD [53]. Nowadays, DL-based CAD can perform automatic nodule detection and automatic Lung-RADS classification. However, it requires radiologists to confirm nodule size, solid part size, and categorization into pure GGN, part-solid nodule, or pure solid nodule step by step before determining the Lung-RADS score. Theoretically, it does not perform Lung-RADS categorization using DL but generates classification results using conditional constructs in its code, without identifying features. For example, in the case of 4X, which refers to category 3 or 4 nodules with additional features or imaging findings that increase the suspicion of malignancy, this classification result relies on the judgment of malignant features. Our model performed better for Lung-RADS scores 2 and 4B + 4X, but is less effective for Lung-RADS scores 3 and 4A. Our model achieved an accuracy of 93.3% in the most severe categorization, 4B + 4X, showing a high degree of consistency with human classification results. The four-class classification had an ordinal severity. This makes it easier to predict the extremes of severity, while the intermediate, ambiguous range is more challenging. Using DL to address the Lung-RADS score classification problem presents several challenges, including the lack of Lung-RADS labeled datasets and the need for a larger dataset for multi-class classification tasks. Ensuring a balanced number of samples for each classification is challenging. As a result, there is relatively little research in this area. In addition, almost all commercial CAD software currently imposes restrictions on nodule sizes, such as 3 mm to 30 mm. However, in our research, we did not limit the nodule size. Therefore, we believe that our model has a broader applicability.
There were some limitations to our study. First, the segmentation of nodules was required for radiomics analysis. Our results showed that radiomics provided additional information apart from that obtained by CNN, indicating that there is potential for improvement in CNN feature extraction. However, with the advent of new tools for automatic or semiautomatic image segmentation, our model could be incorporated into clinical practice in the near future. Second, nodule growth assessment, crucial for Lung-RADS categorization, was not feasible in this study. Our future research will focus on employing AI for the analysis of CT scans obtained at different times to address this issue. Nonetheless, our study provides a promising basis for the development of more accurate and efficient lung nodule classification models.
Conclusion
In conclusion, our proposed model provides a promising approach for accurately classifying pulmonary nodules based on the benign/malignancy, different pathological subtypes, and Lung-RADS system, which could aid in the diagnosis of lung cancer.
Data availability
All datasets generated for this study are included in the article/supplementary material.
Abbreviations
- Lung-RADS:
-
Lung Imaging Reporting and Data System
- LUNA16:
-
Lung Nodule Analysis 2016
- LNOP:
-
Lung Nodule Received Operation
- LNHE:
-
Lung Nodule in Health Examination
- CNN:
-
Convolutional neural network
- NLST:
-
National Lung Screening Trial
- LDCT:
-
Low-dose computed tomography
- MIA:
-
Minimal invasive adenocarcinoma
- IA:
-
Invasive adenocarcinoma
- CAD:
-
Computer-aided detection
- DL:
-
Deep learning
- AIS:
-
Adenocarcinoma in situ
- AAH:
-
Atypical adenomatous hyperplasia
- SqCC:
-
Squamous cell carcinoma
- NAS:
-
Neural architectures search
- CA:
-
Coordinate attention
- CBAM:
-
Convolutional block attention module
- DRDB:
-
Dilated residual dense block
- GLCM:
-
Gray Level Co-occurrence Matrix
- GLRLM:
-
Gray Level Run Length Matrix
- GLSZM:
-
Gray Level Size Zone Matrix
- GLDM:
-
Gray Level Dependence Matrix
- LoG:
-
Laplacian of Gaussian
- SE:
-
Squeeze-and-excitation
- TB:
-
Tuberculosis
- GUI:
-
Graphical user interface
References
Force USPST, Krist AH, Davidson KW, Mangione CM, Barry MJ, Cabana M et al (2021) Screening for lung cancer: us preventive services task force recommendation statement. JAMA 325(10):962–970. https://doi.org/10.1001/jama.2021.1117
Oudkerk M, Liu S, Heuvelmans MA, Walter JE, Field JK (2021) Lung cancer LDCT screening and mortality reduction–evidence, pitfalls and future perspectives. Nat Rev Clin Oncol 18(3):135–151. https://doi.org/10.1038/s41571-020-00432-6
Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5):395–409. https://doi.org/10.1056/NEJMoa1102873
Gao W, Wen CP, Wu A, Welch HG (2022) Association of computed tomographic screening promotion with lung cancer overdiagnosis among asian women. JAMA Intern Med 182(3):283–290. https://doi.org/10.1001/jamainternmed.2021.7769
Meza R, Jeon J, Toumazis I, Ten Haaf K, Cao P, Bastani M et al (2021) Evaluation of the benefits and harms of lung cancer screening with low-dose computed tomography: modeling study for the US preventive services task force. JAMA 325(10):988–997. https://doi.org/10.1001/jama.2021.1077
Lin CY, Chang CC, Huang LT, Chung TJ, Liu YS, Yen YT et al (2021) Computed tomography-guided methylene blue localization: single vs. multiple lung nodules. Front Med (Lausanne) 8:661956. https://doi.org/10.3389/fmed.2021.661956
Mazzone PJ, Lam L (2022) Evaluating the patient with a pulmonary nodule: a review. JAMA 327(3):264–273. https://doi.org/10.1001/jama.2021.24287
de Koning HJ, van der Aalst CM, de Jong PA, Scholten ET, Nackaerts K, Heuvelmans MA et al (2020) Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med 382(6):503–513. https://doi.org/10.1056/NEJMoa1911793
Ost DE, Gould MK (2012) Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med 185(4):363–372. https://doi.org/10.1164/rccm.201104-0679CI
McKee BJ, Regis SM, McKee AB, Flacke S, Wald C (2015) Performance of ACR Lung-RADS in a clinical CT lung screening program. J Am Coll Radiol 12(3):273–276. https://doi.org/10.1016/j.jacr.2014.08.004
Henschke CI, Yip R, Yankelevitz DF, Smith JP (2013) Definition of a positive test result in computed tomography screening for lung cancer: a cohort study. Ann Intern Med 158(4):246–252. https://doi.org/10.7326/0003-4819-158-4-201302190-00004
Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83(5):584–594. https://doi.org/10.4065/83.5.584
Mori M, Rao SK, Popper HH, Cagle PT, Fraire AE (2001) Atypical adenomatous hyperplasia of the lung: a probable forerunner in the development of adenocarcinoma of the lung. Mod Pathol 14(2):72–84. https://doi.org/10.1038/modpathol.3880259
Tsutani Y, Miyata Y, Mimae T, Kushitani K, Takeshima Y, Yoshimura M et al (2013) The prognostic role of pathologic invasive component size, excluding lepidic growth, in stage I lung adenocarcinoma. J Thorac Cardiovasc Surg 146(3):580–585. https://doi.org/10.1016/j.jtcvs.2013.04.032
Borczuk AC, Qian F, Kazeros A, Eleazar J, Assaad A, Sonett JR et al (2009) Invasive size is an independent predictor of survival in pulmonary adenocarcinoma. Am J Surg Pathol 33(3):462–469. https://doi.org/10.1097/PAS.0b013e318190157c
Chiu HY, Chao HS, Chen YM (2022) Application of artificial intelligence in lung cancer. Cancers (Basel) 14(6):1370. https://doi.org/10.3390/cancers14061370
Al Mohammad B, Brennan PC, Mello-Thoms C (2017) A review of lung cancer screening and the role of computer-aided detection. Clin Radiol 72(6):433–442. https://doi.org/10.1016/j.crad.2017.01.002
Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y et al (2017) Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 40:172–183. https://doi.org/10.1016/j.media.2017.06.014
Ciompi F, Chung K, van Riel SJ, Setio AAA, Gerke PK, Jacobs C et al (2017) Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 7:46479. https://doi.org/10.1038/srep46479
Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L et al (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25(6):954–961. https://doi.org/10.1038/s41591-019-0447-x
Tunali I, Gillies RJ, Schabath MB (2021) Application of radiomics and artificial intelligence for lung cancer precision medicine. Cold Spring Harb Perspect Med 11(8):a039537. https://doi.org/10.1101/cshperspect.a039537
Fan L, Fang M, Li Z, Tu W, Wang S, Chen W et al (2019) Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol 29(2):889–897. https://doi.org/10.1007/s00330-018-5530-z
Li D, Mikela Vilmun B, Frederik Carlsen J, Albrecht-Beste E, Ammitzbøl Lauridsen C, Bachmann Nielsen M et al (2019) The performance of deep learning algorithms on automatic pulmonary nodule detection and classification tested on different datasets that are not derived from LIDC-IDRI: a systematic review. Diagnostics (Basel) 9(4):207. https://doi.org/10.3390/diagnostics9040207
Liu X, Hou F, Qin H, Hao A (2018) Multi-view multi-scale CNNs for lung nodule type classification from CT images. Pattern Recogn 77:262–275. https://doi.org/10.1016/j.patcog.2017.12.022
Ashraf SF, Yin K, Meng CX, Wang Q, Wang Q, Pu J et al (2022) Predicting benign, preinvasive, and invasive lung nodules on computed tomography scans using machine learning. J Thorac Cardiovasc Surg 163(4):1496-1505.e1410. https://doi.org/10.1016/j.jtcvs.2021.02.010
Naik A, Edla DR (2021) Lung nodule classification on computed tomography images using deep learning. Wireless Pers Commun 116(1):655–690. https://doi.org/10.1007/s11277-020-07732-1
Wan YL, Wu PW, Huang PC, Tsay PK, Pan KT, Trang NN et al (2020) The use of artificial intelligence in the differentiation of malignant and benign lung nodules on computed tomograms proven by surgical pathology. Cancers (Basel) 12(8):2211. https://doi.org/10.3390/cancers12082211
Tran GS, Nghiem TP, Nguyen VT, Luong CM, Burie JC (2019) Improving accuracy of lung nodule classification using deep learning with focal loss. J Healthc Eng 2019:5156416. https://doi.org/10.1155/2019/5156416
Zhao W, Yang J, Sun Y, Li C, Wu W, Jin L et al (2018) 3D Deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res 78(24):6881–6889. https://doi.org/10.1158/0008-5472.Can-18-0696
Hu Z, Tang J, Wang Z, Zhang K, Zhang L, Sun Q (2018) Deep learning for image-based cancer detection and diagnosis−a survey. Pattern Recogn 83:134–149. https://doi.org/10.1016/j.patcog.2018.05.014
Setio AAA, Traverso A, de Bel T, Berens MSN, Bogaard CVD, Cerello P et al (2017) Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 42:1–13. https://doi.org/10.1016/j.media.2017.06.015
Kim L, Kim KH, Yoon YH, Ryu JS, Choi SJ, Park IS et al (2012) Clinicopathologic and molecular characteristics of lung adenocarcinoma arising in young patients. J Korean Med Sci 27(9):1027–1036. https://doi.org/10.3346/jkms.2012.27.9.1027
Pinsky PF, Berg CD (2012) Applying the national lung screening trial eligibility criteria to the US population: what percent of the population and of incident lung cancers would be covered? J Med Screen 19(3):154–156. https://doi.org/10.1258/jms.2012.012010
Hu Y, Chen G (2015) Pathogenic mechanisms of lung adenocarcinoma in smokers and non-smokers determined by gene expression interrogation. Oncol Lett 10(3):1350–1370. https://doi.org/10.3892/ol.2015.3462
Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91(14):1194–1210. https://doi.org/10.1093/jnci/91.14.1194
Aisner DL, Sholl LM, Berry LD, Rossi MR, Chen H, Fujimoto J et al (2018) The impact of smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations-the lung cancer mutation consortium (LCMC2). Clin Cancer Res 24(5):1038–1047. https://doi.org/10.1158/1078-0432.CCR-17-2289
Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG et al (2011) Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 24(5):653–664. https://doi.org/10.1038/modpathol.2010.232
Liu S, Wang R, Zhang Y, Li Y, Cheng C, Pan Y et al (2016) Precise diagnosis of intraoperative frozen section is an effective method to guide resection strategy for peripheral small-sized lung adenocarcinoma. J Clin Oncol 34(4):307–313. https://doi.org/10.1200/jco.2015.63.4907
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30(9):1234–1248. https://doi.org/10.1016/j.mri.2012.06.010
Shur JD, Doran SJ, Kumar S, Ap Dafydd D, Downey K, O’Connor JPB et al (2021) Radiomics in oncology: a practical guide. Radiographics 41(6):1717–1732. https://doi.org/10.1148/rg.2021210037
Jiang H, Shen F, Gao F, Han W (2021) Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recogn 113:107825. https://doi.org/10.1016/j.patcog.2021.107825
Hou Q, Zhou D, Feng J (2021) Coordinate attention for efficient mobile network design. In: 2021 IEEE/CVF conference on computer vision and pattern recognition (CVPR)
Woo S, Park J, Lee J-Y, Kweon IS (2018) CBAM: convolutional block attention module. Computer vision—ECCV 2018, Cham
Lee H, Matin TN, Gleeson FV, Grau V (2019). Efficient 3D fully convolutional networks for pulmonary lobe segmentation in CT images. ArXiv, abs/1909.07474
Qi K, Wang K, Wang X, Zhang Y, Lin G, Zhang X et al (2023) Lung-PNet: an automated deep learning model for the diagnosis of invasive adenocarcinoma in pure ground-glass nodules on chest CT. AJR Am J Roentgenol. https://doi.org/10.2214/AJR.23.29674
Zhang Y, Feng W, Wu Z, Li W, Tao L, Liu X et al (2023) Deep-learning model of ResNet combined with CBAM for malignant-benign pulmonary nodules classification on computed tomography images. Medicina (Kaunas) 59(6):1088. https://doi.org/10.3390/medicina59061088
Liu G, Liu F, Gu J, Mao X, Xie X, Sang J (2022) An attention-based deep learning network for lung nodule malignancy discrimination. Front Neurosci 16:1106937. https://doi.org/10.3389/fnins.2022.1106937
Marappan S, Mujib MD, Siddiqui AA, Aziz A, Khan S, Singh M (2022) Lightweight deep learning classification model for identifying low-resolution CT images of lung cancer. Comput Intell Neurosci 2022:3836539. https://doi.org/10.1155/2022/3836539
Qi J, Deng Z, Sun G, Qian S, Liu L, Xu B (2022) One-step algorithm for fast-track localization and multi-category classification of histological subtypes in lung cancer. Eur J Radiol 154:110443. https://doi.org/10.1016/j.ejrad.2022.110443
Kao TN, Hsieh MS, Chen LW, Yang CJ, Chuang CC, Chiang XH et al (2022) CT-based radiomic analysis for preoperative prediction of tumor invasiveness in lung adenocarcinoma presenting as pure ground-glass nodule. Cancers (Basel) 14(23):5888. https://doi.org/10.3390/cancers14235888
Park S, Park H, Lee SM, Ahn Y, Kim W, Jung K et al (2022) Application of computer-aided diagnosis for Lung-RADS categorization in CT screening for lung cancer: effect on inter-reader agreement. Eur Radiol 32(2):1054–1064. https://doi.org/10.1007/s00330-021-08202-3
Funding
This work was supported by the National Cheng Kung University Hospital of Taiwan (NCKUH-11201007 and NCKUH-11203049) and the Ministry of Science and Technology of Taiwan (MOST 111-2314-B-006-106).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, CY., Guo, SM., Lien, JJ.J. et al. Combined model integrating deep learning, radiomics, and clinical data to classify lung nodules at chest CT. Radiol med 129, 56–69 (2024). https://doi.org/10.1007/s11547-023-01730-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-023-01730-6