Abstract
The reinvention of potato, from a tetraploid clonal crop into a diploid seed-based hybrid crop, requires insight in the mutational load, recombination landscape, and the genetic basis of fertility. Genomics-based breeding and QTL discovery rely on efficient genotyping strategies such as skim sequencing, to gather genotypic information. The application of skim sequencing to full-sib population of non-inbred parents remains challenging. Here, we report on an R implementation of the OutcrossSeq pipeline for diploids. We applied this pipeline to a large diploid skim sequenced potato population. We used the resulting bin-markers for the construction of high-density parent specific linkage maps, highlighting variation in parental recombination rate and structural variations. We subsequently explored transmission ratio distortion and non-independent assortment of alleles, indicative of large-effect deleterious mutations. Finally, we identified QTLs for seedling tuber yield in pots and pollen shed. This study showcases the range of genetic analyses, from marker inference, identification of transmission ratio distortion, and linkage map construction to QTL mapping, resulting in new insights that contribute to breeding diploid potato.
Similar content being viewed by others
Introduction
Potato (Solanum tuberosum) breeding programmes are undergoing a drastic transformation from the classical tetraploid system and clonal propagation to diploid F1 hybrid breeding system (Lindhout et al. 2011; Jansky et al. 2016). The generation of diploid inbred lines is expected to bolster genetic gains in potato by circumventing the complexity of tetrasomic inheritance and facilitating the fixation of beneficial alleles. The original obstacle of self-incompatibility at the diploid level was recently overcome with the identification of the widespread Sli gene (Clot et al. 2020; Eggers et al. 2021; Ma et al. 2021) or by using dysfunctional S-RNase alleles (Zhang et al. 2019; Enciso-Rodriguez et al. 2019). The second obstacle encountered in this transformation is inbreeding depression. Inbreeding at the diploid level results in the loss of vigour and fertility within a few generations due to the exposure of recessive deleterious mutations. Evaluation of 5377 dihaploids, extracted from tetraploid varieties and progenitor clones, illustrates the problem of mutational load in potato germplasm. Only 60% of these dihaploids set tubers, the majority show problems with flowering and fertility, and only 4% displayed good pollen stainability (Hutten et al. 1995). When the focus on clonal reproduction is replaced by sexual reproduction, fertility issues will prompt for new research projects to understand the genetic factors involved in flowering (Seibert et al. 2020), anther development (Endelman and Jansky 2016), pollen shed, and pollen viability. Despite some effort to identify QTLs regulating self-fertility (Phumichai and Hosaka 2006; Peterson et al. 2016) or pollen stainability (Zhang et al. 2019), the genetic basis of fertility in potato remains elusive. In diploid offspring, large-effect deleterious mutations will result in gametic or zygotic selection and can be identified by studying transmission ratio distortion (TRD) (Zhang et al. 2019). The removal of large-effect deleterious alleles identified via TRD is central to the strategy of hybrid potato genome design proposed by (Zhang et al. 2021). This type of genetic analysis relies on genotyping large segregating populations in a cost-effective way. With the low cost of next-generation sequencing, genotyping-by-sequencing (GBS) strategies are an interesting option. GBS strategies can include a step of genome complexity reduction using restriction enzymes (Elshire et al. 2011), capture baits (Uitdewilligen et al. 2013), or amplicon sequencing like potatoMASH (Leyva-Pérez et al. 2022). Owing to the development of multiplex sequencing (Cronn et al. 2008), methods based on whole genome sequencing at low depth, also known as skim sequencing, have become increasingly popular. However, the low read depth makes robust genotyping challenging for full-sib populations from non-inbred parents. The recently released OutcrossSeq pipeline (Chen et al. 2021) is addressing this issue by local clustering of incomplete sequence variant data to infer genetic markers.
In this study, we implemented the core idea of the OutcrossSeq pipeline for diploids in the R package OutcrossSeqDiploidR and applied it to a skim sequenced diploid potato population of 1536 individuals. We explored the minimal read depth necessary for marker inference and used the resulting markers for the construction of high-density parent specific linkage maps, highlighting variation in parental recombination rate and structural variations on chromosome 3 and 10. Subsequently, we detected nine gametic and one zygotic TRD, as well as four pairs of loci with non-independent assortment of alleles. We identified a major QTL for tuber yield from seedlings in pots co-localising with the position of StCDF1 and 5 minor effect QTLs epistatic to StCDF1 allele combinations. Finally, we identified seven QTLs regulating pollen shed with the largest one located on chromosome 2.
Materials and Methods
Plant Materials
A population of 1536 diploid full-sib seedlings, descending from a cross between two heterozygous potato clones C (USW5337.3) and E (77.2102.37), was sown the 3rd of July 2020. Before transplanting, about one quarter of the seedlings with a crumpled phenotype (Jongedijk et al. 1990) was discarded from the mapping population. A few crumpled seedlings were kept for further analysis of the phenotype. Plants were raised in 19-cm pots in a greenhouse at ambient temperatures (~ 18 °C) and under natural daylight (~ 16–13 h). This population is a backcross (clone C is the female parent of E) with mixed ancestry of Solanum tuberosum Group Tuberosum and Phureja, and S. vernei.
DNA Extraction, Library Preparation, and Sequencing
Young leaf material from the 1536 CE-XW full-sibs were collected on ice in 96 deep well plates and stored at − 20 °C. Genomic DNA extraction was performed at VHL Genetics (Wageningen, NL), and DNA concentration was normalised to 12.5 ng/μl. Sequencing libraries were produced using RipTide DNA library prep kit (iGenomX, Carlsbad, CA) following manufacturer instructions. Briefly, individual samples were labelled in sixteen 96-well plates using well and plate specific barcoded random primers. Once labelled, samples in each plate were pooled together and converted into a NGS library in one single tube. Libraries of each plate were pooled together and sequenced by Limes Innovations B.V (https://limes-innovations.com) with 150-bp paired-end reads using Illumina iSeq 100. The parental clones were sequenced at BaseClear (Leiden, NL) with 100-bp paired-ended reads using Illumina Hiseq2000. The sequencing data generated are available from the European Nucleotide Archive (ENA) under the BioProject ID PRJEB56778.
Demultiplexing, Quality Check, and Alignment
DNA sequences were demultiplexed to individual descendants using the plate and position barcodes with fgbio version 1.4.0 (https://fulcrumgenomics.github.io/fgbio/). Sequence reads of parents and offspring were quality trimmed using fastp version 0.19.5 (Chen et al. 2018) dropping reads with a complexity lower than 20% and a length lower than 70 bp (option-l 70-y 20–5–cut_front_window_size 1–3–cut_tail_window_size 1). Trimmed reads were aligned to the potato reference genome DM v.6.1 using BWA-MEM algorithm v.0.7.17 (Li and Durbin 2009) with default parameters. Alignment summary statistics were extracted with qualimap v.2.2.2 (Okonechnikov et al. 2015).
Variant Calling
Variant calling was performed in the parental clones using bcftools v.1.13 (Danecek et al. 2021) mpileup and call functions and filtered based on quality and depth (option-e ′%QUAL < 20||FORMAT/DP > 40| FORMAT/DP < 10). Parental bcf files were merged, and indels were removed using bcftools view to create a target SNP file with bcftools query (option -f ′%CHROM\t%POS\t%REF,%ALT\n′). This target file was used to constrain variant calling in each offspring sample to these target coordinates and alleles using bcftools call -T and -C options. Variant calling in the offspring was parallelised by chromosome. Multiallelic sites were split into multiple rows using bcftools norm –m, and the resulting chromosomal bcf files were converted into chromosomal variant matrices, filtering out variants with a missing rate above 0.5 using VCFtools v0.1.16 (Danecek et al. 2011) (options–max-missing 0.50–012).
Clustering of Adjacent Sequence Variants into Robust Marker Genotypes with OutcrossSeqDiploidR
The R package OutcrossSeqDiploidR (https://github.com/ccrclot/OutcrossSeqDiploidR) was used to cluster offspring, which co-inherited the same heterozygous and private sequence variants of either parent in windows of 0.1 Mb. Based on cluster membership, genotypes were inferred resulting in potentially 7312 intervals of 0.1 Mb for each parent hereafter referred to as ‘bin-markers’. Our R package follows the core idea of the OutcrossSeq pipeline for diploids (Chen et al. 2021), relying on a reference genome to locally infer genotypes from a population kinship matrix. However, in this implementation, female and male variants are clustered independently to estimate uniparental genotypes rather than the integrated genotyping information produced by the original pipeline. In addition, a quality check function was implemented allowing users to flag bin-markers with aberrant recombination or transmission ratio values. Recombination percentages above 50% allowed to bring all (initially randomly assigned) bin-markers alleles from the same parental chromosome into linkage phase. All steps are summarised in Fig. 1 and in the package vignette. Firstly, the function recodeParentMatList was used to (1) recode variant dosage into presence absence data, since artefactual homozygous absence could be derived from low coverage information on a heterozygous site, and (2) extract heterozygous variant privates to each parent (1 × 0 and 0 × 1) into separate female and male specific binary matrix. The following steps of OutcrossSeqR pipeline were applied independently to chromosomal female and male matrices. The function makeBins with parameter bin_size = 100,000 was used to group variants in physical bins of 100 kb. Within each bin, the Euclidian distance between individuals was calculated based on presence absence of variants with the function getDist. The resulting kinship matrix was then used to cluster the individuals using the ward.D2 method with the function clusterPop. The function getHap with parameter k = 2 was used to extract, from the clustering data, two groups of individuals per bin-marker, corresponding to the two alleles that could be inherited form one parent. The function qualCheck with default parameters was then used to flag noisy bin-markers displaying (1) high recombination rates with adjacent bins (0.3 < r < 0.7) and (2) deviations exceeding 0.15 points from the local transmission ratio, calculated with five flanking markers at either side. Flagged bin-markers were removed. Bin-marker data with r > 0.5 were swapped to phase adjacent bin-markers with the function phaseHap. Phased bin-markers were visualised, and phasing was manually improved, when necessary, with the function manualPhasing before being formatted for Smooth Descent (Navarro et al. 2022) with the function formatSD.
Correcting Genotyping Errors with Smooth Descent
The segregation data of bin-markers obtained with OutcrossSeqDiploidR were corrected for putatively erroneous data points with the algorithm Smooth Decent following the package vignette (Thérèse Navarro et al. 2023). Briefly, Smooth Decent makes use of identity-by-descent probabilities, in our case based on the physical order of bin-markers, to detect putative genotyping errors and impute the most probable genotype given the data points of flanking markers. A total of seven iterative rounds of data improvement with Smooth Descent were performed. The first five rounds were based on a prediction interval of 1 Mb and the two final rounds on a prediction interval of 5 and 10 Mb.
Linkage Map Construction
The cleaned bin-marker data were used to generate independent male and female linkage maps with polymapR version 1.1.2 (Bourke et al. 2018) following the package vignette. Recombination frequencies were calculated for all pairs of bin-markers. Bin-markers were assigned to 12 chromosomal linkage groups of either parent, based on their physical positions on DM v6.1 reference genome. Next, bin-markers were ordered using MDSmap_from_list, a wrapper function around the estimate.map function from MDSMap (Preedy and Hackett 2016). During the mapping process, 47 outlying bin-markers with a high nearest-neighbour fit score or an abnormal position in the principal curve analysis were removed.
Chromosomal Recombination Rate and Marey Map
The chromosomal recombination rate (cM/Mb) was calculated using the genetic length of a linkage map and the physical length of the corresponding chromosomes of DM v6.1 assembly. Similarly, Marey maps (Chakravarti 1991) were derived from the comparison between the physical and genetic position of each marker. Finally, the positions of CENH3-binding domains on DM v6.1 were obtained from Pham et al. (2020).
Transmission Ratio Distortion and Non-independent Assortment of Alleles
Male and female bin-markers were tested for significant deviation from the expected Mendelian 1:1 ratio via a χ2 test with 1 degree of freedom and a p value for significance set at 0.01. Following the method used by Dukić and Bomblies (2022), we corrected for multiple testing in 23 chromosome arms and utilised a significance threshold of α = 0.01/23 ≈ 0.0004 (χ2 = 12.4, df = 1). A region with transmission ratio distortion (TRD) was considered biologically meaningful when markers displayed significant TRD over at least 2 Mb. Using the same threshold for significance, TRD due to zygotic rather than gametic selection were detected with a χ2 test based on a 2 × 2 contingency table where the observed frequencies of one of the four possible haplotype combinations were compared with the expected frequencies based on random assortment of the observed parental haplotypes frequencies.
During the construction of the linkage map, we detected associations of bin-markers alleles located on different linkage groups. We decided to explore those deviations from independent assortment of alleles between all marker pairs using the G-test option implemented in the function linkage of ploymapR. Like the zygotic χ2 test, this option calculates the G2 statistic for independence based on a 2 × 2 contingency. Instead of returning a G2 value, this function returned a LOD score of independence and defined as \(\frac{{G}^{2}}{\mathrm{log}\left(100\right)}\). We considered that bin-markers pairs with a LOD of independence > 5 were displaying biologically significant deviations from independent assortment.
Phenotyping Seedling Yield in Pots and Pollen Shed
Phenotypic data on flowering and pollen shed was collected from the seventh to the tenth week of the growing season. The ability to produce at least one flower reaching anthesis was recorded and resulted in a binary classification. A pollen sample (extracted with an electric toothbrush from a freshly open flower at anthesis) was collected for each flowering seedling. If the pollen extraction of the first collected flower failed, the plant was resampled. When only a few pollen grains could be extracted after resampling of four flowers on two different days, the plant was classified as a poor pollen shedder, resulting in a binary classification for pollen shed (poor vs. good). After a growing cycle of 18 weeks, three quarters of the population had senesced, and tuber harvest started. Over a period of 2 weeks, tubers larger than 5 mm were harvested in one paper bag per descendant. The fresh weight in gram was measured with an electronic scale and used as yield per pot estimate.
QTL Mapping
QTL mapping was performed using the package polyqtlR version 0.0.6 (Bourke et al. 2021). The function singleMarkerRegression was used to fit an additive model at each marker position returning the − log10 p value of model fit per marker. The significance thresholds for QTL detection were determined via permutation tests on the phenotypic values with N = 1000 cycles and α = 0.05. QTL discovery was based on the separate maternal and paternal maps, but at significant QTL positions, we analysed the phenotypic effects of the combined male and female haplotypes. This was done with a one-way ANOVA with multi-comparison Tukey’s HSD post hoc test (α = 0.05). To estimate the effects size of QTLs and their potential interactions, all significant QTLs, and their interactions if significant, were used as factor in (generalised) linear models predicting for the phenotype of interest. While a linear regression was used for tuber yield, we used a logistic regression for the binary trait pollen shed and estimated effect size with odds ratio.
Results
Analysis of Sequencing Data
The sequencing of the libraries comprising the 1536 seedlings yielded 3364 gigabases (Gb) which after demultiplexing resulted in 2910 Gb. After trimming and quality filtering, we obtained 19,766,020,170 usable reads corresponding to a total of 2562 Gb, which corresponds to an average of 1.7 Gb per seedling. A proportion of 93.6% of reads could be mapped to DM v6.1 generating a median coverage per individual of 1.34 × , or 0.67 × per haploid genome (Suppl. Figure 1). A total of 18 Gb and 22.6 Gb were obtained for the parental clone C and E. After trimming and quality filtering, we obtained 149,677,752 and 182,224,166 usable reads corresponding to a total of 14.8 Gb and 18 Gb. A proportion of 98.7% and 98.6% of reads could be mapped to DM v6.1 generating a coverage of 18.72 × for clone C and 22.47 × for clone E or 9.36 × and 11.24 × per haploid genome. After variant calling and quality filtering in the parents, we obtained a total of 12,983,013 variants which were subsequently called in the offspring. Only, a subset of 1,889,749 variant could be observed in more than 50% of the offspring. Knowing that the low read-depth will yield erroneous dosage information, we focused on heterozygous parent-private variant (1 × 0 and 0 × 1). A total of 554,806 and 902,714 were heterozygous and private to clones C and E, respectively. The distribution of heterozygous private variants was not uniform across parents and chromosomes (Fig. 2a). For example, on chromosome 7, the female parent C showed a fivefold higher number of heterozygous parent-private variants than the male parent E. For chromosomes 3, 6, and 11, the reverse was observed, where the male parent was more polymorphic (Fig. 2b). Regions where parents differ in variant density tend to coincide with the location of the pericentromeric heterochromatin (Suppl. Figure 2). We concluded that linkage map construction based on segregating sequence variants was not possible. The low sequencing depth disallows dosage estimates to distinguish between homozygous or heterozygous offspring. The presence of heterozygous parent-private variants can be scored, but their absence can be bona fide (due to inheritance) or the result of low coverage information. This motivated us to use the presence/absence information of all co-inherited heterozygous parent-private variants from a 0.1-Mb interval to infer the genotype score of this interval as a so-called bin-marker. As a result, we generated sparser but more robust genotyping data.
Distribution of heterozygous parent-private sequence variant. a Total number of parent-private variant per chromosome per parent. b 5 Mb moving average of the number of heterozygous parent-private variants across bins of 100 kb. Clone C data are displayed in orange and clone E in blue. The triangles indicate regions with a low density of heterozygous parent-private variants disallowing the inference of bin-markers
Inference of Bin-Markers with OutcrossSeqDiploidR
The DM v6.1 reference genome was subdivided in 7312 bins of 100 kb to infer bin-markers. The parent-private variants within these bins were used to cluster descendants and to infer the segregating alleles of each bin-marker. Regions with a low density of parent-private variants, in combination with the low sequencing depth, lacked information to infer bin-marker genotypes. This resulted in larger chromosomal intervals without genetic markers. The regions with low density of parent-private variants are plotted in Fig. 2b, and the impact on bin-marker imputation and thus genetic map coverage is shown in Table 1 and Fig. 3. We arrived at 7145 female and 7174 male bin-markers, which had at least one heterozygous parent-private variant. After all subsequent cleaning steps (see the “Materials and Methods” section), we obtained 4935 female and 4746 female bin-markers with genetically and biologically sound characteristics. Although this represented a ~ 34% reduction of markers, this predominantly affected marker coverage at pericentromeric regions.
Smooth Descent Improved Genotyping Data
Smooth Descent detected and imputed 1,075,417 erroneous datapoints, corresponding to 7.2% of our bin-marker dataset. Data inspection showed that Smooth Descent was unable to clean noise in several plants because of a too unfavourable signal to noise ratio due to low sequencing depths. To identify a threshold to reject such plants, we compared sequence coverage with the amount of noise. The expected number of true genetic recombination events per plant is low. One erroneous data point suggests two extra recombination events relative to the flanking markers. When counting all ‘recombination events’ per plant from a noisy dataset, we reached values, shown in Suppl. Figure 3, so much inflated by error (exceeding 500), that we can use the number of ‘recombination events’ to indicate noise. Unsurprisingly, the amount of recombination (noise) was not randomly distributed across individuals but increased with decreasing genome coverage. Smooth Descent corrected more erroneous datapoints in plants with low coverage, but this ability reached a tipping point and started to decrease around an average diploid coverage of 0.3 × (Suppl. Figure 3). The 75 individuals with a coverage below 0.3 × were removed. The remaining 1461 individuals were used in subsequent analyses.
High-Density Linkage Maps
After filtering 47 outlying bin-markers during the mapping process, a total of 4894 male and 4740 female bin-markers segregating across 1461 individuals could be mapped on 24 linkage groups. Those linkage groups corresponded to the 12 paternal and 12 maternal chromosomes (Table 1). On the female side, the average number of bin-markers per linkage group was 408 ranging from 257 for chromosome 11 to 549 for chromosome 1. On the male side, the average of 395 bin-markers per linkage group was comparable, with a minimum of 99 bin-markers for chromosome 7 and a maximum of 523 for chromosome 9. The marker density and gaps (up to 16.9 cM) in the genetic maps corresponded to chromosomal regions with a low density of parent-private variants (Fig. 2b). In addition, extreme transmission ratio distortion (TRD) also hampered to infer bin-markers, resulting in gaps on female and male chromosome 1 of 6.4 and 8.9 cM, respectively. Nonetheless, both parental maps were of high density with an average interval between bin-markers of 0.3 cM on the female and 0.2 cM on the male maps.
Structural Variation and Chromosomal Recombination Rate Variation
Comparing the physical bin-marker positions, using DM v6.1 reference genome, with the genetic marker positions on parent specific linkage maps, we could estimate an average genome wide recombination rate of 1.58 cM/Mb for the female clone C and of 1.07 cM/Mb for the male clone E, and this genome-wide difference was also true for every chromosome (Table 2). In both parental maps, the lowest chromosomal recombination rate per Mb was observed on chromosome 10 and the highest one on the acrocentric chromosome 2 with values ranging from 1.33 to 2.02 cM/Mb for clone C and 0.78 to 1.29 for clone E. As expected, the recombination rate was far from uniform along each chromosome with virtually no recombination in pericentromeric regions as evidenced by the horizontal sections around the positions of CENH3-binding domains, indicated with triangles, on the Marey maps (Fig. 2). In addition, two euchromatic regions without recombination were observed in clone C. On the south arm of chromosome 3, a segment of 5.8 Mb from 42.9 to 48.7 Mb was found, and a 6.3 Mb segment from 52.8 to 59.1 Mb on chromosome 10. In clone E, this chromosome 3 segment was inverted relatively to DM suggesting the presence of a homozygous inversion. In clone C, this inversion must be heterozygous, explaining the suppression of recombination. In addition, the north arms of chromosomes 3 and 6 of clone E displayed a strong reduction of recombination.
Transmission Ratio Distortion
We detected significant gametic TRD (α = 0.0004) at loci located on eight different chromosomes (Fig. 4). On the female map four loci displayed significant TRD. The locus with the largest deviation (~ 1:6) was identified on chromosome 1 (distortion clone C chromosome 1, abbreviated as DC1), at 69.65 Mb. Loci with milder TRD were identified on chromosomes 4 (DC4 at 58.65 Mb) and 12 (DC12 at 9.35 Mb). A locus with weak but significant TRD was also detected in the pericentromeric region of chromosome 5. On the male map, six loci displayed significant TRD. Again, the locus with the largest deviation was detected on chromosome 1 (DE1), but its localisation in or near the pericentromeric heterochromatin is inaccurate due to the inability to infer bin-markers. On chromosome 11, another extreme TRD locus (~ 9:1) was mapped at 6.55 Mb (DE11). Except for the last 10 Mb, the entire chromosome 8 showed mild TDR. This could be due to the presence of two TRD loci at either side of the pericentromeric region as suggested by the mild local maxima observed on this chromosome (DE8a at 4.45 Mb and DE8b at 44.55 Mb). Finally, significant but weak TRD regions were identified on chromosome 2 (32.05 to 34.45 Mb), the short arm of chromosome 4 and the peri-centromeric regions of chromosome 9. The loci with strong TDR coincide with previously described loci such as the S-locus and ar1 for DE1 and DC1 on chromosome 1 and la1 for chromosome 8 (Jacobs et al. 1995; Zhang et al. 2019; see the “Discussion” section). We then investigated if those TRD could be caused by zygotic rather than gametic selection. Only one case of zygotic selection was observed on chromosome 1 at locus DZ1 (distortion zygotic chromosome 1) located at 69.65 Mb. DZ1 co-locates with DC1 at 69.65 Mb on the female map, but the localisation of the male counterpart is blurry because the gametic selection in the pericentromeric region (DE1) adds to the zygotic selection around 69.65 Mb (Suppl. Figure 4). We postulate that the near absence of one male by female allele combination is the result of selection against about one quarter of the seedlings with the crumpled phenotype (Suppl. File 2), which were excluded beforehand from analysis.
Non-independent Assortment of Alleles
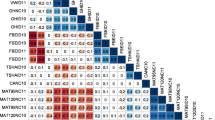
Following the detection of a zygotic TRD, resulting in non-independent assortment of alleles at one locus, we investigated deviations from independent assortment between all bin-marker pairs and observed biologically significant deviations (LOD of independence > 5) on three female and five male chromosomes. As expected, male and female bin-marker alleles around 69.6 to 70.7 Mb on chromosome 1 displayed extreme deviation from independent assortment due to the zygotic TRD DZ1 (Fig. 5b). A mild deviation from independent assortment was identified between marker pairs from female chromosomes 4 and 12 around the position of DC4 and DC12 (Fig. 5c). Another association of comparable significance was identified between most of male chromosome 3 (from the low recombining north arm to the south arm inversion) and the pericentromere of chromosome 9 already associated with DE9 (Fig. 5d). Finally, two regions near DE11 on male chromosome 11 showed a highly significant deviation from independent assortment of alleles: one from 5.3 to 6.6 Mb interacting with most of male chromosome 3 and the other from 7.6 to 8.2 Mb interacting with the DE8a region on male chromosome 8 (Fig. 5e). While close, the LOD-2 confidence interval of those two regions on chromosome 11 did not overlap suggesting the presence of two separate, albeit linked loci showing these associations.
a Overview of pairs of loci showing non-independent assortment of alleles. Ribbons link non-independently associated chromosomal region (LOD-2 confidence interval). Pair-wise non-independent assortments occurring between parents are depicted in green; the one occurring within parent is depicted in orange for C and in blue for E. b–e Heatmap displaying the LOD of independence of chromosomes with biological significant non-independent assortment of alleles (LOD > 5) against physical position
QTL Mapping for Seedling Yield in Pots
Phenotypic data for tuber yield in pots, defined as fresh weight of tubers larger than 5 mm, were collected on 1534 clones after a growing cycle of 18 weeks. Tuber yield ranged from 0 g (no tubers above 5 mm) to 213.1 g with a mean value of 69.1 g (Fig. 6a). Three significant QTLs were identified for tuber yield (TY): the minor QTL TYE8 (QTL for tuber yield clone E chromosome 8) and the major QTLs TYC5 and TYE5 (Table 3, Fig. 6b). The LOD-1.5 confidence interval around the top marker of TYC5 and TYE5 overlap with the position of the candidate gene StCDF1. In this population, alleles StCDF1.1 and StCDF1.2 are known to segregate from clone C and alleles StCDF1.1 and StCDF1.3 from clone E (Kloosterman et al. 2013). Interestingly, the significance for QTL discovery differs vastly between parents (TYE5 − log10(p value) = 118.7; TYC5 − log10(p value) = 23.7), indicating a larger phenotypic effect on tuber yield by the StCDF1.3 allele. This observation is better understood when the effects on tuber yield of the four StCDF1 haplotype combinations are compared (Fig. 6c). Both StCDF1.2 and StDCF1.3 haplotypes contributed to TY, but with unequal effect (Tukey’s HSD post hoc test α = 005). The large-effect StCDF1.3 haplotype was dominant over the minor-effect StCDF1.2 haplotype. The deviation from additivity was highly significant (p < 0.001). Overall, in this population, 44.7% of variation in tuber yield can be explained by variation in haplotype combination at the StCDF1 locus.
a Distribution of tuber yield (TY) (n = 1459). b Localisation of significant QTLs for tuber yield on the male and female map of chromosome 5. The X-axis represents physical position (Mb), the Y-axis represents − log10(p value), and threshold of significance is indicated by the black dashed line. Clone C data are displayed in orange and clone E in blue. The triangle indicates the position of StCDF1 on DM v6.1. c Raincloud plots illustrating the effect of StCDF1 haplotype combinations on TY. The different letters and colours indicate haplotype combinations with significantly different tuber yield (Tukey’s HSD post hoc test α = 0.05)
To cancel the overwhelming effect of the StCDF1 locus, we performed QTL discovery for tuber yield within three subpopulations with equal maturity, corresponding to cohorts of plants homozygous for StCDF1.1, the StCDF1.1 / StCDF1.2 heterozygotes, and offspring with the StCDF1.3 allele. Now, five minor-effect QTLs surfaced, two in the low yielding StCDF1.1 homozygotes, one in the medium yielding 1.2/1.1 cohort, and two in the high yielding cohort with StCDF1.3 (Table 3; Suppl. Figure 5). Interestingly, each sub-population resulted in new QTL positions. For example, QTL TYC1 explained 5% of tuber yield variation in the 1.2/1.1 cohort but did not account for a significant difference in the other two cohorts. Similarly, clone C chromosome 7 bears on each side of its centromere QTLs TYC7a and TYC7b specific to the 1.1/1.1 and -/1.3 cohort, respectively. These observations showed that tuber yield was not only controlled by StCDF1 allelic combinations, but that these allelic combinations also epistatically controlled how minor QTLs can exert their effects.
QTL Mapping for Pollen Shed
During the growing season, most plants flowered profusely, except 57 offspring plants that did not flower or dropped their buds. Using the ability to produce flowers as binary trait, we could not identify any significant QTL regulating flower production. Out of the remaining 1479 flowering plants, 134 were classified as poor pollen producers and 1345 as good pollen producers. Using this binary classification as a phenotype, we identified seven significant QTLs regulating pollen shed (Fig. 6a, Table 4). PSE2 (QTL pollen shed clone E chromosome 2), a QTL with a LOD score of 14.6, was identified on clone E chromosome 2 located at around 37.65 Mb. QTLs with LOD scores ranging from 4.5 to 7.7 were identified on C chromosomes 1, 5, 10, and 11 and on E chromosomes 1 and 9 and are detailed in Table 4. The associations between pollen shed and segregating marker alleles are presented as contingency tables visualised as mosaic plots in Fig. 6b. Holding the effect of other QTLs constant, the odds of being a low pollen shedder were 8.1 times higher for the plants bearing the detrimental allele at PSE2. We therefore considered PSE2 as a major QTL controlling pollen shed in this population (Fig. 7).
a Location of significant QTLs for pollen shed (n = 1406). The X-axis represents physical position (Mb), the Y-axis represents − log10(p value), and threshold of significance is indicated by the black dashed line. Clone C data are displayed in orange and clone E in blue. b Mosaic plots illustrate the effect of the different haplotypes on the poor pollen shedder’s classification at QTL positions. Counts of poor pollen shedders are displayed in red and counts of good pollen shedders in green
Discussion
OutcrossSeqDiploidR Bin-Markers Inference
This study explored the construction of linkage maps of skim sequenced offspring at a very low depth. This resulted in elevated missing values at SNP loci which prohibited linkage analysis. However, grouping of parent-private SNPs in windows of 0.1 Mb allowed clustering of descendants to impute bin-markers. Performance of OutcrossSeqDiploidR was hampered at specific regions with either high or low sequence divergence. High sequence divergence of the parents relative to the DM reference genome results in failure of read mapping. Low sequence divergence among the parental alleles resulting in less than ~ 30 parent-private variant per bin of 100 kb also excluded accurate imputation of bin-markers. Such regions were observed on maternal (clone C) chromosomes 3, 6, and 11 and on paternal (clone E) chromosome 7. Bin-markers from such incongruent or nearly invariant regions were recognised as inconsistent and removed by subsequent quality filters. The loss of one-third of the potential number of bin-markers may have little impact on our results, because most markers were lost in the non-recombining heterochromatic regions and potentially in truly homozygous regions. However, we occasionally noticed a small detrimental effect on the precision of QTL mapping when bin-markers were lost in regions of absolute TRD. Contrary to the original OutcrossSeq pipeline, we did not choose to replace the flagged bin-makers with values of adjacent bin-markers, so it remains clear to users where genotyping failed.
In this study, we explored the minimal depth of skim sequencing to infer genetically consistent bin-markers of 0.1 Mb. We used the sum of recombination events between physically adjacent marker-bins as a metric to monitor the performance of Smooth Descent to correct spurious datapoints. We determined that our R package could not generate genetically consistent bin-markers data when the diploid genome coverage was below 0.3 × . This agrees with the test results of the diploid outcrossing module of the original OutcrossSeq pipeline which also showed increasing error rates ranging from 1.29 to 14.05% for decreasing coverage ranging from 4 × to 0.5 × . While agreeing with Chen et al. (2021) who recommend a minimum coverage of 1 × to ensure sufficient genotyping power, we show that in a diploid potato population of 1536 individuals, a coverage lower than 1 × but higher than 0.3 × can still be used for 100 kb bin-marker inference by OutcrossSeqDiploiR, in combination with the algorithm Smooth Decent to correct genotyping errors.
Comparison of Genetic and Physical Maps
Bin-markers were used to construct separate maternal and paternal linkage maps. The quality and specific features of these maps were evaluated by comparing the genetic and physical bin-marker position with Marey maps. This allowed us to visualise an approximately 5.8 Mb paracentric inversion on the long arm of E chromosome 3 (42.9–48.7 Mb) relative to the DM reference genome. While parent E is homozygous for this inversion, parent C must be heterozygous, because the same ~ 5.8 Mb interval did not show recombination events. The same inversion was recently reported in a diploid potato pan-genome study and validated by chromatin interaction (Tang et al. 2022). This inversion appears to be rather common and was observed in 12 out of 20 S. tuberosum diploid landraces. A second region without recombination of 6.3 Mb length, observed on the linkage map of clone C of chromosome 10, could be identical to the inversion reported on chromosome 10 from 52.7 to 59.1 Mb in S. tuberosum Group Andigenum clone PG6244 (i.e., CIP 703,509). Genetic map length, reflecting the recombination frequency, of the maternal clone C is always higher than the one of the paternal clone E. This agrees with Anithakumari et al. (2010) who reported genetic maps length of 1012.4 cM for clone C and 774.6 for clone E. We cannot determine if this difference in recombination rate is due to heterochiasmy or other factors because the reciprocal cross never succeeded.
Transmission Ratio Distortion
Loci subject to TRD have been reported in almost every mapping study in diploid potato (Manrique-Carpintero et al. 2016). The relevance of their identification to avoid inbreeding depression was recently highlighted (Zhang et al. 2019, 2021). We reported three female, six male loci with gametic TDR and one locus showing zygotic selection. Here, the underlying causes will be discussed. TRD locus DE1, located in the pericentromeric region of the paternal chromosome 1 is due to the S-locus involved in gametophytic self-incompatibility (Gebhardt et al. 1991; Jacobs et al. 1995). The zygotic TRD at locus DZ1 centred around at 69.65 Mb on chromosome 1 in our population coincides with the ar1 locus and candidate gene StSIEL identified in clone E (i.e., PG2662) selfing population by Zhang et al. (2019). Their phenotypic description of ar1 mutants, abnormal rooting and shoot differentiation, partially agrees with the phenotype crumpled, as previously reported in CxE population by Jongedijk et al. (1990) and detailed in Suppl. File 2. More generally, all of the paternal TRD observed in our population coincide with TRD presented by Zhang et al. (2019) in E selfing population. For instance, the gametic TRD at locus DE8a is identical to the locus la1. Here, we show that in addition to the previously reported zygotic selection against homozygous la1, the la1/DE8a region is also subject to gametic selection. Furthermore, the strong gametic TRD at locus DE11 is identical to an unnamed but equally strong zygotic TRD reported by the same authors. Those potentially conflicting observation on the gametic or zygotic stage of TRD could be reconciled assuming that clone C is homozygous for the allele causing zygotic TRD. In addition, most of the TRD loci identified in maternal clone C overlap with previously identified TRD in selfing population of S. tuberosum Group Phureja clones E86-69, C10-20, and PG6359 (Zhang et al. 2021). However, overlapping TRD regions identified in different clones can be due to a shared locus or to independent loci located in the same region. The TRD at locus DC12 overlaps with a TRD region observed in clone PG6359 selfings. The TRD at locus DC4 overlaps with TRD regions observed in clone E86-69 and C10-20 selfings that could not be linked with a visible phenotypic defect. In addition to TRD, we explored non-independent assortment of alleles between pairs of loci and to our knowledge are the first to report their presence in a potato population. Most of the associated pairs of loci overlap with regions displaying TRD such as DE8a and DE11 or DC4 and DC12. Interestingly, this was not the case for the low recombining male chromosome 3 involved in non-independent assortments of alleles with both chromosome 9 and 11 without harbouring TRD. Non-independent assortment of alleles between pairs of unliked loci can be explained by the presence of co-adapted gene complexes (Clegg et al. 1972). Alternatively, such associations can be found in interspecific crosses where they can be caused by Bateson–Dobzhansky–Muller incompatibilities (Bateson 1909; Dobzhansky 1936; Muller 1942) resulting in selection against heterospecific allele combinations in hybrids. Some of the associations observed in our population could be due to such incompatibilities since the parental clones of our population present a mixed ancestry of S. tuberosum Group Tuberosum and Phureja, and S. vernei. Overall, the identification of unliked loci having alleles that positively or negatively associate with each other adds a new layer of complexity to the understanding of inbreeding depression in potato. When designing the genome of inbred potato, as proposed by Zhang et al. (2021), one should consider that complementation and interaction are not only allelic phenomena but also inter-locus ones, acting between different genomic regions.
Seedling Tuber Yield in Pots
The identification of the StCDF1 locus as the major-effect QTL regulating yield in pots in our population is consistent with previous mapping studies (Manrique-Carpintero et al. 2015; Marand et al. 2019) and with the molecular function of StCDF1, which has been described as the master regulator of potato maturity (Kloosterman et al. 2013). Maturity is observed by comparing the status of above-ground plant development (foliage discolouration, prostrated stems, end of apical sprout, and flowering activity) to the maturity status of reference varieties. The maturity values predict the duration of the life cycle of field grown cultivars, which varies between 80 and 140 days. Maturity also predicts the dependency on daylength to trigger tuber development. The wild-type allele StCDF1.1 is commonly present in genotypes with a prolonged growing season and dependency on short-day conditions for tuberization. StCDF1.2 and StCDF1.3 alleles are found in early maturing genotypes adapted to long-day conditions. Breeders are exploiting the dosage dependent effect of StCDF1 alleles on maturity to breed cultivars adapted to different growing condition and market segments. For example, the very late maturing starch cultivars Altus and Avenger are quadruplex for StCDF1.1 (Hoopes et al. 2022) resulting in a prolonged growing season which maximise accumulation of starch. While long growing seasons are usually associated with higher yields in cultivars, our pot-grown seedlings show that early genotypes, bearing StCDF1.2 or StCDF1.3 haplotypes yielded more than late genotypes homozygous for StCDF1.1. This counter-intuitive outcome can be explained by the delayed tuberization of genotypes homozygous for StCDF1.1. Our 1.1/1.1 cohort with delayed tuberization is genetically different in comparison to commercial varieties, which are selected for early tuber set, independent of maturity. Interestingly, in our population, the positive effect of StCDF1.3 on tuber yield was larger than the one of StCDF1.2. Similarly, a stronger effect of StCDF1.3 over StCDF1.2 on early maturity was also estimated in the offspring the tetraploid cultivars Altus and Columba (Hoopes et al. 2022). Those results are consistent with the finding that StFLORE, the long non-coding RNA and antisense transcript of StCDF1, is disrupted in StCDF1.3 but not in StCDF1.2 (Ramírez Gonzales et al. 2021). Disruption of StFLORE makes StCDF1.3 heterozygotes theoretically less susceptible to translation inhibition than StCDF1.2 heterozygotes. While the effect of maturity on yield is usually controlled with a co-factor, our large population allowed us to perform independent QTL discovery within three maturity cohorts and unravel minor effect QTLs specific to a given maturity class. With this approach, we identified five minor effect QTLs, all of them specific to a single cohort, suggesting GxG interaction between StCDF1 allele combinations and those minor-effect QTLs. Given the epistatic effect of StCDF1 allele combinations over the minor effect QTLs identified, we argue that deciphering the genetic component(s) of yield should be performed in fixed StCDF1 background. For example, non-StCDF1 factors contributing to early tuberization might substantially contribute to yield in late maturing genotypes and must have been selected for in cultivars such as Altus and Avenger (Hoopes et al. 2022), but their effects will be masked in early maturing genotypes bearing StCDF1.2 or StCDF1.3 alleles. Because of the pleiotropic effect of maturity on starch content which later influence frying colour and cooking type, this argument may also hold for other traits.
Candidate Genes for Pollen Shed
While poor male fertility is a historical issue in potato breeding (Krantz 1924), the ongoing re-invention of potato into a diploid F1 hybrid crop (Lindhout et al. 2011; Jansky et al. 2016) is turning this trait into a major breeding target. Fertility starts with the ability to produce flowers for which we could not find QTL in our population. Male fertility is subsequently determined by the ability to produce and release pollen for which we identified one major QTL PPE2 on chromosome 2 with a top bin-marker located at 37.65 Mb and a LOD-1.5 confidence interval ranging from 33.6 to 38.1 Mb. Studying TRD at the fruiting stage followed by a bulked segregant analysis for floral bud abortion, Zhang et al. (2021) identified StDYT1 (Soltu.DM.02G019340) as a regulator of stamen development and thus affecting fertility. StDYT1 is located approximately at 33.6 Mb on the chromosome 2 of DMv6.1 assembly, right at the boundary of the LOD-1.5 confidence interval of QppE2, and could be considered as a candidate gene. We identify two other candidate genes involved in anther development more closely located to the top bin-marker of QppE2: Soltu.DM.02G024440 and Soltu.DM.02G023940. Soltu.DM.02G024440 is homologous to the S. lycopersicum transcription factor Tomato MADS box gene 6 (TM6). TM6 RNAi lines and TM6 deletions mutant ms-15 display homeotic defects primarily in stamen and are male sterile (de Martino et al. 2006; Cao et al. 2019). Since carpelloid stamens were only observed on a few clones out of the entire population, it is unlikely that a loss-of-function allele of Soltu.DM.02G024440 is segregating our population. However, altered expression level of Soltu.DM.02G024440 resulting in aberrant anthers development and subsequent poor pollen shed cannot be excluded. The second candidate gene, Soltu.DM.02G023940, is homologous to the A. thaliana homeobox transcription factor WUSCHEL (WUS). WUS is regulating stem cell activity in the organising centre of floral meristem and is implicated in floral patterning (Ikeda et al. 2009). In addition, Deyhle et al. (2007) reported the involvement of WUS in anther development more particularly in the differentiation of stomium cells. During normal development, stomium cells undergo cell wall thickening and degenerate which leads to rupture of the anther and pollen release. In wus mutants, this process is repressed resulting in indehiscent anthers and no pollen release which resemble our poor pollen shed phenotype.
Data Availability
The sequencing data are available from the ENA under the BioProject ID PRJEB56778. The software OutcrossSeqDiploidR is available at https://github.com/ccrclot/OutcrossSeqDiploidR. The phenotyping data, genotyping data, and all the R code to reproduce the results and figures of this article are available at https://figshare.com/s/1d07942fe2b7518974f0.
References
Anithakumari AM, Tang J, van Eck HJ et al (2010) A pipeline for high throughput detection and mapping of SNPs from EST databases. Mol Breed 26:65–75. https://doi.org/10.1007/s11032-009-9377-5
Bateson W (1909) Heredity and variation in modern lights. Darwin and modern science
Bourke PM, van Geest G, Voorrips RE et al (2018) polymapR—linkage analysis and genetic map construction from F1 populations of outcrossing polyploids. Bioinformatics 34:3496–3502. https://doi.org/10.1093/bioinformatics/bty371
Bourke PM, Voorrips RE, Hackett CA et al (2021) Detecting quantitative trait loci and exploring chromosomal pairing in autopolyploids using polyqtlR. Bioinformatics 37:3822–3829. https://doi.org/10.1093/bioinformatics/btab574
Cao X, Liu X, Wang X et al (2019) B-class MADS-box TM6 is a candidate gene for tomato male sterile-15 26. Theor Appl Genet 132:2125–2135. https://doi.org/10.1007/s00122-019-03342-z
Chakravarti A (1991) A graphical representation of genetic and physical maps: the Marey map. Genomics 11:219–222. https://doi.org/10.1016/0888-7543(91)90123-V
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen M, Fan W, Ji F et al (2021) Genome-wide identification of agronomically important genes in outcrossing crops using OutcrossSeq. Mol Plant 14:556–570. https://doi.org/10.1016/j.molp.2021.01.003
Clegg MT, Allard RW, Kahler AL (1972) Is the gene the unit of selection? Evidence from two experimental plant populations. Proc Natl Acad Sci USA 69:2474–2478. https://doi.org/10.1073/pnas.69.9.2474
Clot CR, Polzer C, Prodhomme C et al (2020) The origin and widespread occurrence of Sli-based self-compatibility in potato. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03627-8
Cronn R, Liston A, Parks M et al (2008) Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res 36:122. https://doi.org/10.1093/nar/gkn502
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Danecek P, Bonfield JK, Liddle J et al (2021) Twelve years of SAMtools and BCFtools. GigaScience 10:1–4. https://doi.org/10.1093/gigascience/giab008
de Martino G, Pan I, Emmanuel E et al (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18:1833–1845. https://doi.org/10.1105/tpc.106.042978
Deyhle F, Sarkar AK, Tucker EJ, Laux T (2007) WUSCHEL regulates cell differentiation during anther development. Dev Biol 302:154–159. https://doi.org/10.1016/j.ydbio.2006.09.013
Dobzhansky TH (1936) Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21:113
Dukić M, Bomblies K (2022) Male and female recombination landscapes of diploid Arabidopsis arenosa. Genetics 220. https://doi.org/10.1093/genetics/iyab236
Eggers EJ, van der Burgt A, van Heusden SAW et al (2021) Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat Commun 12:1–9. https://doi.org/10.1038/s41467-021-24267-6
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS one 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Enciso-Rodriguez F, Manrique-Carpintero NC, Nadakuduti SS et al (2019) Overcoming self-incompatibility in diploid potato using CRISPR-Cas9. Front Plant Sci 10:376. https://doi.org/10.3389/fpls.2019.00376
Endelman JB, Jansky SH (2016) Genetic mapping with an inbred line-derived F2 population in potato. Theor Appl Genet 129:935–943. https://doi.org/10.1007/s00122-016-2673-7
Gebhardt C, Ritter E, Barone A et al (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor Appl Genet 83:49–57. https://doi.org/10.1007/BF00229225
Hoopes G, Meng X, Hamilton JP et al (2022) Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Mol Plant 15:520–536. https://doi.org/10.1016/j.molp.2022.01.003
Hutten RCB, Soppe WJJ, Hermsen JGT, Jacobsen E (1995) Evaluation of dihaploid populations from potato varieties and breeding lines. Potato Res 38:77–86. https://doi.org/10.1007/BF02358072
Ikeda M, Mitsuda N, Ohme-Takagi M (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an Activator in floral patterning. Plant Cell 21:3493–3505. https://doi.org/10.1105/tpc.109.069997
Jacobs JME, Van Eck HJ, Arens P et al (1995) A genetic map of potato (Solanum tuberosum) integrating molecular markers, including transposons, and classical markers. Theor Appl Genet 91:289–300. https://doi.org/10.1007/BF00220891
Jansky SH, Charkowski AO, Douches DS et al (2016) Reinventing potato as a diploid inbred line-based crop. Crop Sci 56:1412–1422. https://doi.org/10.2135/cropsci2015.12.0740
Jongedijk E, van der Wolk JMASA, Suurs LCJM (1990) Analysis of glutamate oxaloacetate transaminase (GOT) isozyme variants in diploid tuberous Solanum; inheritance and linkage relationships to ds1 (desynapsis), y (tuber flesh colour), cr (crumpled) and yc (yellow cotyledon). Euphytica 45:155–167. https://doi.org/10.1007/BF00033282
Kloosterman B, Abelenda JA, Gomez MDMC et al (2013) Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495:246–250. https://doi.org/10.1038/nature11912
Krantz FA (1924) Potato breeding methods. Technical Bulletin 25:
Leyva-Perez M de la O, Vexler L, Byrne S et al (2022) PotatoMASH—a low cost, genome-scanning marker system for use in potato genomics and genetics applications. Agronomy 12:2461. https://doi.org/10.3390/agronomy12102461
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Lindhout P, Meijer D, Schotte T et al (2011) Towards F1 hybrid seed potato breeding. Potato Res 54:301–312. https://doi.org/10.1007/s11540-011-9196-z
Ma L, Zhang C, Zhang B et al (2021) A nonS-locus F-box gene breaks self-incompatibility in diploid potatoes. Nat Commun 12:1–8. https://doi.org/10.1038/s41467-021-24266-7
Manrique-Carpintero NC, Coombs JJ, Cui Y et al (2015) Genetic map and QTL analysis of agronomic traits in a diploid potato population using single nucleotide polymorphism markers. Crop Sci 55:2566–2579. https://doi.org/10.2135/cropsci2014.10.0745
Manrique-Carpintero NC, Coombs JJ, Veilleux RE et al (2016) Comparative analysis of regions with distorted segregation in three diploid populations of potato. G3: Genes Genomes Genet 6:2617–2628. https://doi.org/10.1534/g3.116.030031
Marand AP, Jansky SH, Gage JL et al (2019) Residual heterozygosity and epistatic interactions underlie the complex genetic architecture of yield in diploid potato. Genetics 212:317–332. https://doi.org/10.1534/genetics.119.302036
Muller H (1942) Isolating mechanisms, evolution, and temperature. In: Biol. Symp. 71–125
Navarro AT, Bourke PM, van de Weg E et al (2022) Smooth Descent: a ploidy-aware algorithm to improve linkage mapping in the presence of genotyping errors
Okonechnikov K, Conesa A, García-Alcalde F (2015) Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:btv566. https://doi.org/10.1093/bioinformatics/btv566
Peterson BA, Holt SH, Laimbeer FPE et al (2016) Self-fertility in a cultivated diploid potato population examined with the infinium 8303 potato single-nucleotide polymorphism array. Plant Genome 9:10. https://doi.org/10.3835/plantgenome2016.01.0003
Pham GM, Hamilton JP, Wood JC et al (2020) Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience 9:1–11. https://doi.org/10.1093/gigascience/giaa100
Phumichai C, Hosaka K (2006) Cryptic improvement for fertility by continuous selfing of diploid potatoes using Sli gene. Euphytica 149:251–258. https://doi.org/10.1007/s10681-005-9072-5
Preedy KF, Hackett CA (2016) A rapid marker ordering approach for high-density genetic linkage maps in experimental autotetraploid populations using multidimensional scaling. Theor Appl Genet 129:2117–2132. https://doi.org/10.1007/s00122-016-2761-8
Ramírez Gonzales L, Shi L, Bergonzi SB, Oortwijn M, Franco-Zorrilla JM, Solano-Tavira R, Visser RGF, Abelenda JA, Bachem CW (2021) Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J 105(4):855–869
Seibert T, Abel C, Wahl V (2020) Flowering time and the identification of floral marker genes in Solanum tuberosum ssp. andigena. J Exp Bot 71:986–996. https://doi.org/10.1093/jxb/erz484
Tang D, Jia Y, Zhang J et al (2022) Genome evolution and diversity of wild and cultivated potatoes. Nature 2022:1–7. https://doi.org/10.1038/s41586-022-04822-x
Thérèse Navarro A, Bourke PM, van de Weg E et al (2023) Smooth Descent: a ploidy-aware algorithm to improve linkage mapping in the presence of genotyping errors. Front Genet 14
Uitdewilligen JGAML, Wolters A-MA, D’hoop BB et al (2013) A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8:e62355. https://doi.org/10.1371/journal.pone.0062355
Zhang C, Wang P, Tang D et al (2019) The genetic basis of inbreeding depression in potato. Nat Genet 51:374–378. https://doi.org/10.1038/s41588-018-0319-1
Zhang C, Yang Z, Tang D et al (2021) Genome design of hybrid potato. Cell. https://doi.org/10.1016/j.cell.2021.06.006
Acknowledgements
We thank the members of the public-private partnership “A new method for potato breeding: the ‘Fixation-Restitution’ approach” and SusCrop ERANET funded project “DIFFUGAT: Diploid Inbreds For Fixation, and Unreduced Gametes for Tetraploidy” (Averis Seeds B.V., Bejo Zaden B.V., Danespo A/S, Germicopa, Den Hartigh B.V., SaKa Pflanzenzucht GmbH & Co. KG, C. Meijer B.V., and Teagasc) for providing their support. We are grateful to Christian W.B. Bachem, Sara Bergonzi and, Li Shi for their input on StCDF1 discussion. Our colleagues of Unifarm are acknowledged for plant care in the greenhouse.
Funding
The projects “A new method for potato breeding: the ‘Fixation-Restitution’ approach” and “DIFFUGAT: Diploid Inbreds For Fixation, and Unreduced Gametes for Tetraploidy” were respectively financially supported by the Dutch Topsector Horticulture & Starting Materials (grant number TU18075) and SusCrop ERANET (grant number 106). Within the Topsector, private industry, knowledge institutes, and the government are working together on innovations for sustainable production of safe and healthy food and the development of a healthy green environment.
Author information
Authors and Affiliations
Contributions
CRC: conceived the research, collected and analysed phenotypical data, constructed libraries, implemented OutcrossSeqDiploidR, performed bioinformatic and genetic analysis, and wrote the manuscript. XW: collected and analysed phenotypical data and constructed libraries. JK: collected and analysed phenotypical data. ATN: contributed to Smooth Descent results. JB: constructed libraries. RGFV: supervised research and reviewed manuscript. RF: supervised research and reviewed manuscript. HJvE: obtained funding, conceived and supervised research, and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
R.G.F. Visser and H.J. van Eck are members of the editorial board of Potato Research. The other authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Clot, C.R., Wang, X., Koopman, J. et al. High-Density Linkage Map Constructed from a Skim Sequenced Diploid Potato Population Reveals Transmission Distortion and QTLs for Tuber Yield and Pollen Shed. Potato Res. 67, 139–163 (2024). https://doi.org/10.1007/s11540-023-09627-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11540-023-09627-7