Abstract
Key message
The paper proposes and validates a robust method for rapid construction of high-density linkage maps suitable for autotetraploid species.
Abstract
Modern genotyping techniques are producing increasingly high numbers of genetic markers that can be scored in experimental populations of plants and animals. Ordering these markers to form a reliable linkage map is computationally challenging. There is a wide literature on this topic, but most has focussed on populations derived from diploid, homozygous parents. The challenge of ordering markers in an autotetraploid population has received little attention, and there is currently no method that runs sufficiently rapidly to investigate the effects of omitting problematic markers on map order in larger datasets. Here, we have explored the use of multidimensional scaling (MDS) to order markers from a cross between autotetraploid parents, using simulated data with 74–152 markers on a linkage group and also experimental data from a potato population. We compared different functions of the recombination fraction and LOD score to form the MDS stress function and found that an LOD2 weighting generally performed well, including when missing values and genotyping errors are present. We conclude that an initial analysis using unconstrained MDS gives a rapid method to detect and remove problematic markers, and that a subsequent analysis using either constrained MDS or principal curve analysis gives reliable marker orders. The latter approach is also particularly rapid, taking less than 10 s on a set of 258 markers compared to 6 days for the JoinMap software. This MDS approach could also be applied to experimental populations of diploid species.




Similar content being viewed by others
References
Cheema J, Dicks J (2009) Computational approaches and software tools for genetic linkage map estimation in plants. Brief Bioinform 10:595–608
Cheema J, Ellis NTH, Dicks J (2010) THREaD Mapper Studio: a novel, visual web server for the estimation of genetic linkage maps. Nucleic Acids Res 38:W188–W193
de Leeuw J, Mair P (2009) Multidimensional scaling using majorization: SMACOF in R. J Stat Softw 31:1–30
Elshire RJ, Glaubitz JC, Qi S, Poland JA, Kawamoto K, Buckler E, Mitchell SE (2011) A robust, simple Genotyping by Sequencing (GbS) approach for high diversity species. PLoS One 6(5):e19379. doi:10.1371/journal.pone.0019379
Felcher KJ, Coombs JJ, Massa AN, Hansey CN, Hamilton JP et al (2012) Integration of two diploid potato linkage maps with the potato genome sequence. PLoS One 7:e36347. doi:10.1371/journal.pone.0036347
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Hackett CA, Pande B, Bryan GJ (2003) Constructing linkage maps in autotetraploid species using simulated annealing. Theor Appl Genet 106:1107–1115
Hackett CA, Milne I, Bradshaw JE, Luo ZW (2007) TetraploidMap for Windows: linkage map construction and QTL mapping in autotetraploid species. J Hered 98:727–729
Hackett CA, McLean K, Bryan GJ (2013) Linkage analysis and QTL mapping using SNP dosage data in a tetraploid potato mapping population. PLoS One 8:e63939
Haldane JBS (1919) The combination of linkage values, and the calculation of distances between the loci of linked factors. J Genet 8:299–309
Hastie T, Stuetzle W (1989) Principal curves. J Am Stat Assoc 84:502–516
Hastie T, Weingessel A (2013) Princurve: fits a principal curve in arbitrary dimension. R package version 1.1-12. http://CRAN.R-project.org/package=princurve
Lalouel JM (1977) Linkage mapping from pair-wise recombination data. Heredity 38:61–77
Liu BH (1998) Statistical genomics. CRC Press, Boca Raton
Luo ZW, Hackett CA, Bradshaw JE, McNicol JW, Milbourne DM (2001) Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157:1369–1385
Maliepaard C, Jansen J, Van Ooijen JW (1997) Linkage analysis in a full-sib family of an outbreeding plant species: overview and consequences for applications. Genet Res 67:55–65
Newell WR, Mott R, Beck S, Lehrach H (1995) Construction of genetic maps using distance geometry. Genomics 30:59–70
Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–197
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
Rastas P, Paulin L, Hanski I, Lehtonen R, Auvinen P (2013) Lep-Map: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29:3128–3134
Schiex T, Gaspin C (1997) CarthaGène: constructing and joining maximum likelihood genetic maps. In: Proceedings of the fifth international conference on intelligent systems for molecular biology, vol 97, pp 258–267
Sharma SK, Bolser D, de Boer J, Sonderkaer M, Amoros W, Carboni MF, D’Ambrosio JM, de la Cruz G, Di Genova A, Douches DS, Equiluz M, Guo X, Guzman F, Hackett CA, Hamilton JP, Li G, Li Y, Lozano R, Maass A, Marshall D, Martinez D, McLean K, Mejia N, Milne L, Munive S, Nagy I, Ponce O, Ramirez M, Simon R, Thomson SJ, Torres Y, Waugh R, Zhang Z, Huang S, Visser RGF, Bachem CWB, Sagredo B, Feingold SE, Orjeda G, Veilleux RE, Bonierbale M, Jacobs JME, Milbourne D, Martin DMA, Bryan GJ (2013) Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3 Genes Genom Genet 3:2031–2047
Shields DC, Collins A, Buetow KH, Morton NE (1991) Error filtration, interference and the human linkage map. Proc Natl Acad Sci USA 88:6501–6505
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JOINMAP. Plant J 3:739–744
Stam P, Van Ooijen JW (1995) JoinMap™ version 2.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen
Van Ooijen JW (2006) JoinMap® 4; software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen
Van Ooijen JW, Jansen J (2013) Genetic mapping in experimental populations. Cambridge University Press, Cambridge
Van Os H, Stam P, Visser RG, van Eck HJ (2005) RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112:30–40
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wu Y, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from minimum spanning tree of a graph. PLoS Genet 4(10):e1000212. doi:10.1371/journal.pgen.1000212
Zhao H, Speed TP (1996) On genetic map functions. Genetics 142:1369–1377
Acknowledgments
The financial support for this work from the Scottish Government’s Rural and Environment Science and Analytical Services Division (RESAS) is gratefully acknowledged. We thank Dr. Glenn Bryan, Dr. Karen McLean and colleagues at the James Hutton Institute for use of the potato genotype data and information on the potato reference sequence, and Dr. Herman van Eck and the anonymous reviewers for their constructive comments during the revision of this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by H. J. van Eck.
Electronic supplementary material
Below is the link to the electronic supplementary material.
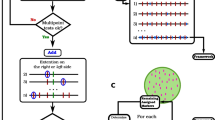
Appendix: Details of the MDS algorithms
Appendix: Details of the MDS algorithms
Principal curves MDS
-
1.
Use the smacofSym function from the R package smacof (1.4-0) (de Leeuw and Mair 2009) to perform two- or three-dimensional weighted unconstrained MDS on the distance matrix.
-
2.
Plot the final configuration to find potential outliers from Smacofsym plot (see Fig. 1 solid circles for a two-dimensional example and Fig. 3 for a three-dimensional example)
-
3.
Fit the principal curves using the method of Hastie and Stuetzle (1989) implemented in the R package princurve (version 1.1-12) (Hastie and Weingessel 2013).
-
4.
Plot the first principal curve on the final configuration of the unconstrained fit and assess whether it looks reasonable.
-
5.
The projections of the markers onto the first principal curve give the estimated map positions.
Constrained MDS
Steps 1–2 as for principal curve
-
3.
Use the smacofSphere function in two dimensions to constrain the points to approximate to the arc of a circle with a penalty, p, for deviations from the arc.
-
4.
Plot the final configuration from smacofSym and smacofSphere to check for any points which have major changes in rank with respect to either dimension in the final configuration (Supplementary Figure 1A).
-
5.
Check the stress ratio smacofsphere stress/smacofsym stress. This is a metric for the increase in stress (which approximates to a measure of the reduction in fit) caused by forcing the points to lie on an arc and should be below 1.1. If the ratio is above this, return to step 4 and reduce the penalty p.
-
6.
Project the final configuration onto a line to get order and estimated map length.
-
(a)
Centre sphere on (0, 0).
-
(b)
Calculate the polar coordinates of each point in the configuration.
-
(c)
Rotate, so that the mapping starts at the beginning of the arc.
-
(d)
Radius of the sphere is the median distance of points from (0, 0) rescaled, so that the sum of the configuration is the same as the sum of the observed distances. (We also considered using the mean distance, but this made little difference and the median is less sensitive to outliers and so results are not presented here.)
-
(e)
Order the markers by increasing the angle.
-
(f)
Intermarker distances are equal to the radius multiplied by the difference in angle between the points.
-
(a)
Rights and permissions
About this article
Cite this article
Preedy, K.F., Hackett, C.A. A rapid marker ordering approach for high-density genetic linkage maps in experimental autotetraploid populations using multidimensional scaling. Theor Appl Genet 129, 2117–2132 (2016). https://doi.org/10.1007/s00122-016-2761-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2761-8