Abstract
A gene regulatory network summarizes the interactions between a set of genes and regulatory gene products. These interactions include transcriptional regulation, protein activity regulation, and regulation of the transport of proteins between cellular compartments. DSGRN is a network modeling approach that builds on traditions of discrete-time Boolean models and continuous-time switching system models. When all interactions are transcriptional, DSGRN uses a combinatorial approximation to describe the entire range of dynamics that is compatible with network structure. Here we present an extension of the DGSRN approach to transport regulation across a boundary between compartments, such as a cellular membrane. We illustrate our approach by searching a model of the p53-Mdm2 network for the potential to admit two experimentally observed distinct stable periodic cycles.









Similar content being viewed by others
Code Availability
Any code used in this work is available upon request.
References
Abou-Jaoude W, Ouattara DA, Kaufman M (2009) From structure to dynamics: frequency tuning in the p53-mdm2 network i. Logical approach. J Theor Biol 258:561–577
Abou-Jaude W, Chaves M, Gouze J-L (2011) A theoretical exploration of birhythmicity in the p53-mdm2 network. PLoS ONE 6(2):e17075
Albert R, Collins JJ, Glass L (2013) Introduction to focus issue: quantitative approaches to genetic networks. Chaos 23(2):025001
Barak Y, Juven T, Haffner R, Oren M (1993) Mdm2 expression is induced by wild type p53 activity. EMBO 12:461–468
Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol. Call 21:307–315
Chen J, Lin J, Levine AJ (1995) Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142–152
Crawford-Kahrl P, Cummins B, Gedeon T (2021) Joint realizability of monotone boolean functions. under review
Cummins B, Gedeon T, Harker S, Mischaikow K (2017) Database of dynamic signatures generated by regulatory networks (dsgrn). In Jerome F, Heinz K (eds) Computational Methods in Systems Biology, 2017, chapter 19, Springer, pp 300–308
Cummins B, Gedeon T, Harker S, Mischaikow K, Mok K (2016) Combinatorial representation of parameter space for switching systems. SIAM J Appl Dyn Syst 15(4):2176–2212
Cummins B, Gedeon T, Harker S, Mischaikow K (2018) Model rejection and parameter reduction via time series. SIAM J Appl Dyn Syst 17(2):1589–1616
Cummins C, Gameiro M, Gedeon T, Kepley S, Mischaikow K, Zhang L (2021) Extending combinatorial regulatory network modeling to include activity control and decay modulation. arXiv:2111.01399 [math.DS]
de Jong H (2002) Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol 9:67–103
De Jong H, Gouzé JL, Hernandez C, Page M, Sari T, Geiselmann J (2004) Qualitative simulation of genetic regulatory networks using piecewise-linear models. Bull Math Biol 66(2):301–340, 3
Duncan T, Gedeon W (2021) Stability and bifurcations of equilibria in networks with piecewise linear interactions. Int J Bif Chaos 31(11):2130032
Duncan T, Gedeon W, Kokubu H, Mischaikow K, Oka H (2021) Equilibria and their stability in networks with steep sigmoidal nonlinearities
Edwards R (2001) Chaos in neural and gene networks with hard switching. Diff Eq Dyn Syst 9:187–220
Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a ring finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275:8945–8951
Fillipov AF (1988) Differential equations with discontinuous righthand side. Kluwer Academic Publishing, Dordrecht
Freedman DA, Wu L, Levine AJ (1999) Functions of the mdm2 oncoprotein. Cell Mol Life Sci 55:96–107
Gameiro M, Gedeon T, Kepley S, Mischaikow K (2021) Rational design of complex phenotype via network models. PLoS Comp Biol 17(7):e1009189
Gatz SA, Wiesmuller L (2006) p53 in recombination and repair. Cell Death Differ 13(6):1003–1016
Gedeon T, Harker S, Kokubu H, Mischaikow K, Oka H (2017) Global dynamics for steep sigmoidal nonlinearities in two dimensions. Physica D 339:18–38
Gedeon T (2020) Multi-parameter exploration of dynamics of regulatory networks. BioSystems 190:1045113
Gedeon T, Cummins B, Harker S, Mischaikow K (2018) Identifying robust hysteresis in networks. PLOS Comput Biol 14(4):1–23, 04
Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A et al (2006) Oscillations and variability in the p53 system. Mol Syst Biol. https://doi.org/10.1038/msb4100068
Glass L, Kauffman S a (1972) Co-operative components, spatial localization and oscillatory cellular dynamics. J Theor Biol 34(2):219–37
Glass L, Kauffman SA (1973) The logical analysis of continuous, non-linear biochemical control networks. J Theor Biol 39(1):103–29
Gottlieb TM, Leal JFM, Seger R, Taya Y, Oren M (2002) Cross-talk between akt, p53 and mdm2: possible implications for the regulation of apoptosis. Oncogene 21:1299–1303
Inoue T, Geyer RK, Howard D, Yu ZK, Maki CG (2001) Mdm2 can promote the ubiquitination, nuclear export, and degradation of p53 in the absence of direct binding. J Biol Chem 276:45255–45260
Ironi I, Panzeri L (2009) A computational framework for qualitative simulation of nonlinear dynamical models of gene-regulatory networks. BMC Bioinform 10:S14
Ironi L, Panzeri L, Plahte E, Simoncini V (2011) Dynamics of actively regulated gene networks. Physica D Nonlinear Phenom 240(8):779–794
Jeffrey MR (2018) Hidden Dynamics: The Mathematics of Switches. Springer, Decisions and Other Discontinuous Behaviour
Kepley S, Mischaikow K, Zhang L (2021) Computing linear extensions for polynomial posets subject to algebraic constraints. SIAM J Appl Algebra Geom 5(2):388–416
Kruse J-P, Gu W (2009) Modes of p53 regulation. Cell 137:609–622
Machina A, Edwards R, van den Driessche P (2013) Sensitive dependence on initial conditions in gene networks. Chaos 23:025101
Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW et al (1993) Oncoprotein mdm2 conceals the activation domain of tumour suppressor p53. Nature 362:857–860
Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10(4):431–442
Ouattara DA, Abou-Jaude W, Kaufman M (2010) From structure to dynamics: frequency tuning in the p53-mdm2 network. ii differential and stochastic approaches. J Theor Biol 264:1177–1189
Plahte E, Kjoglum S (2005) Analysis and generic properties of gene regulatory networks with graded response functions. Physica D 201:150–176
Plahte E, Mestl T, Omholt SW (1994) Global analysis of steady points for systems of differential equations with sigmoid interactions. Dyn Stabil Syst 9(4):275–291
Thomas R (1991) Regulatory networks seen as asynchronous automata: a logical description. J Theor Biol 153:1–23
Veflingstad SR, Plahte E (2007) Analysis of gene regulatory network models with graded and binary transcriptional responses. Biosystems 90(2):323–339
Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408:307–310
Xin Y, Cummins B, Gedeon T (2020) Multistability in the epithelial-mesenchymal transition network. BMC Bioinform 21(1):24
Acknowledgements
The work of B.C. and T. G. was partially supported by NSF grant DMS-1839299 and grant DARPA FA8750-17-C-0054. The work of W.D and T.G. was additionally supported by an grant NIH 5R01GM126555-01. We acknowledge the Indigenous nations and peoples who are the traditional owners and caretakers of the land on which this work was undertaken at the Montana State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no known conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A Representative ODE Simulations
Appendix A Representative ODE Simulations
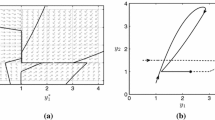
Here we present representative simulations of continuous ODE system (15) in Figs. 10 and 11. Both figures show simulations at a parameter sampled from the parameter node \(M_1^F\), the parameter node with a middle black wall with the front unfolding. The parameter used in Fig. 10 results in a large attracting periodic orbit, while the parameter in Fig. 11 results in a small attracting periodic orbit. The parameters used in each figure are given in Table 7.
Representative large periodic orbit simulation. A and B: Projections of solutions to (15) onto the Mc-Mn and Mc-P planes, respectively. Trajectories are shown for 10 different initial conditions. After a brief transient, all trajectories converge to the periodic orbit. C–E: Time series plots for one of the trajectories shown in (A) and (B). The orbit is large because P crosses both of its thresholds
Representative small periodic orbit simulation. A and B: Projections of solutions to (15) onto the Mc-Mn and Mc-P planes, respectively. After a brief transient, all orbits converge to the periodic orbit. C–E: Time series plots for one of the trajectories shown in (A) and (B). The orbit is small because P crosses only one of its thresholds
Rights and permissions
About this article
Cite this article
Fox, E., Cummins, B., Duncan, W. et al. Modeling Transport Regulation in Gene Regulatory Networks. Bull Math Biol 84, 89 (2022). https://doi.org/10.1007/s11538-022-01035-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-022-01035-1