Abstract
Background
Sasanlimab is a monoclonal antibody that binds to the programmed cell death receptor 1 (PD-1). Anti-PD-1 monoclonal antibodies have improved patient clinical outcomes; however, not all treated patients derive clinical benefit. Further insights on potential biomarkers beyond PD-L1 expression levels would help to identify the patients most likely to respond to treatment.
Objective
This study evaluated tumor biopsies from patients treated with intravenous or subcutaneous sasanlimab to identify biomarkers of response and characterize pharmacodynamic activity.
Methods
Anti-PD-1/PD-ligand 1 (PD-L1)-naive patients with advanced solid tumors received sasanlimab intravenously at 1, 3, or 10 mg/kg every 3 weeks (n = 23) or subcutaneously at 300 mg every 4 weeks (n = 15). Best tumor percentage change from baseline was determined by RECIST. Whole-exome DNA and RNA sequencing were performed in tumor samples collected from treated patients at protocol-defined timepoints. PD-L1 and CD8 protein expression were evaluated in tumor biopsies by immunohistochemistry. Associations with response were assessed by linear regression analysis.
Results
Baseline tumor mutational burden (TMB), as well as PD-L1 and CD8 expression, were significantly associated with response to sasanlimab across the multiple dose levels, routes of administration, and range of tumor types evaluated. TMB is an independent biomarker from the various tumor inflammatory genes and signatures evaluated. Gene set enrichment analysis showed that higher baseline expression levels of genes related to the interferon-γ and PD-1 signaling pathways and the cell cycle were significantly associated with response to sasanlimab across tumor types. No differences were observed between routes of administration with regard to response to sasanlimab for the biomarkers of interest (TMB, PD-L1, CD8, and interferon-γ signature). Evaluation of pharmacodynamic changes showed increased tumor expression of genes enriched in adaptive immune response pathways.
Conclusions
Our findings indicate an active, immunomodulatory mechanism for the anti-PD-1 antibody sasanlimab across different tumor types and routes of administration.
Trial Registration
ClinicalTrials.gov identifier NCT02573259; registered October 2015.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This is the first study evaluating biomarkers and pharmacodynamic changes with both intravenous and subcutaneous routes of administration of the anti-PD-1 monoclonal antibody, sasanlimab. |
An increased expression of adaptive immune activation genes/pathways in on-treatment biopsies suggests an active, immunomodulatory mechanism for sasanlimab therapy across different tumor types and routes of administration. |
1 Introduction
Anti-programmed cell death receptor 1 (PD-1) or anti-PD ligand 1 (PD-L1) monoclonal antibodies improve patient clinical outcomes, with long-lasting responses and prolonged survival across a wide range of advanced malignancies [1,2,3,4,5,6,7,8,9]. However, not all treated patients derive clinical benefit. Further insights on potential biomarkers are needed beyond PD-L1 expression levels, to identify the patient populations most likely to respond to treatment [2, 10,11,12].
Sasanlimab (PF-06801591) is a humanized, immunoglobulin G4 monoclonal antibody that binds to the PD-1 receptor with high affinity and blocks its interaction with PD-1 ligands [13,14,15]. Clinical findings from the first-in-human, dose-escalation, phase I study (B8011001) of sasanlimab administered intravenously (IV) or subcutaneously (SC) in patients with locally advanced or metastatic solid tumors showed that treatment was well tolerated at all the investigated dose levels (0.5–10 mg/kg IV every 3 weeks (q3w) and 300 mg SC every 4 weeks (q4w)) [14, 15]. Full PD-1 receptor occupancy in peripheral CD8+ T cells was also observed at all dose levels, independent of baseline PD-1 expression levels [15]. Responses were achieved in patients with ovarian cancer, sarcoma, small-cell lung cancer (SCLC), squamous cell carcinoma of the head and neck (SCCHN), and endometrial adenocarcinoma with high microsatellite instability, following IV or SC treatment [15].
In the current study, tumor biopsies available from patients treated with sasanlimab administered IV or SC, across a wide range of advanced solid tumor types, were evaluated to identify biomarkers of response and pharmacodynamic effects of sasanlimab, independent of cancer histology and route of administration.
2 Methods
2.1 Study Design and Treatment
This was a phase I, open-label, multicenter, dose-escalation (Part 1) and dose-expansion (Part 2) study conducted in patients with locally advanced or metastatic solid tumors. The primary endpoint of Part 1 was to evaluate safety and tolerability of treatment with sasanlimab administered IV or SC. Antitumor clinical activity (objective response rate) was assessed as a secondary endpoint; evaluations of pharmacodynamic changes and biomarkers were included as exploratory study objectives.
In dose escalation, patients were treated with sasanlimab IV at 0.5, 1, 3, or 10 mg/kg q3w or SC 300 mg q4w according to the modified toxicity probability interval method [16]. Additional patients were enrolled into the 1 mg/kg IV, 3 mg/kg IV, 10 mg/kg IV, and 300 mg SC cohorts for further pharmacodynamic assessments. Treatment with sasanlimab continued until disease progression, unacceptable toxicity, or withdrawal of consent.
2.2 Patients
Patients were eligible for enrollment if they had melanoma, ovarian cancer, sarcoma, SCCHN, NSCLC, urothelial cancer, or other advanced solid tumor types known to respond to anti-PD-1 or anti-PD-L1 therapy, but not previously treated with anti-PD-1 or anti-PD-L1 therapy. In addition, patients were required to have measurable disease and disease progression on at least one prior line of treatment or have refused standard-of-care therapy. Further patient inclusion and exclusion criteria were as previously described [15]. The flow of samples collected from patients during the study is shown in Fig. 1.
The study was approved by the institutional review board or independent ethics committee of the participating institutions and followed the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice guidelines. All patients provided written informed consent. The study was sponsored by Pfizer and was registered at ClinicalTrials.gov (NCT02573259).
2.3 Assessment of Clinical Antitumor Activity
Tumor responses were evaluated by computed tomography or magnetic resonance imaging at baseline and then every 6 weeks (IV group) or every 8 weeks (SC group) until disease progression, death, subsequent anticancer therapy, withdrawal of consent, or at the end of treatment, using the Response Evaluation Criteria in Solid Tumors (RECIST) v1.1 [17].
2.4 Whole-Exome Sequencing (WES) and RNA-Seq
Tumor biopsy samples were collected at baseline and on day 8 of cycle 2 (C2D8) from patients treated in Part 1 at 1, 3, and 10 mg/kg IV and 300 mg SC in the safety and pharmacodynamics cohorts (archival samples from the safety cohorts; baseline and C2D8 samples from the pharmacodynamics cohorts, Fig. 1). DNA and RNA sequencing of formalin-fixed, paraffin-embedded tumor samples were conducted at Personalis Inc. (Menlo Park, CA, USA). Exome capture was performed using Agilent SureSelect Clinical Research Exome according to the manufacturers’ recommendations. Additional supplementation with Personalis ACE proprietary target probes was performed to enhance coverage in difficult-to-sequence regions within sets of biomedically and clinically relevant genes, as previously described [18]. Briefly, manufacturer protocols were modified to adjust the average library insert length to ~ 250 base pairs (bp) and the use of KAPA HiFi DNA Polymerase for DNA sequencing and stranded RNA Sequencing kit for RNA sequencing (Kapa Biosystems, Wilmington, MA, USA). Sequencing was performed on NovaSeq 600 (Illumina, San Diego, CA, USA) sequencers with paired-end 2 × 150 bp read lengths and Illumina’s proprietary reversible terminator-based method. For DNA sequencing, tumor specimens were sequenced to an average depth of coverage of 200× across the 69.4 Mb ACE assay genomic footprint. For RNA sequencing, tumor specimens were sequenced to an average output of 50 million paired end reads (total of 100 M reads). Tumor samples were further analyzed through Personalis somatic DNA pipeline for small variant calling (single nucleotide variants, InDels), and calculation of the tumor mutational burden (TMB). TMB, defined as non-synonymous somatic variants/Mb, was estimated from WES. For RNA sequencing, tumor samples were analyzed through Personalis RNA pipeline for gene expression data.
Twenty-seven baseline tumor samples were evaluated for TMB. Best overall response (BOR) was determined by RECIST (data cutoff: 19 April 2019) for 24 of the 27 patients with baseline tumor samples: six of these patients achieved a partial response (PR), eight had stable disease (SD), and ten experienced progressive disease (PD).
RNA-Seq was performed on 40 tumor samples (26 baseline and 14 C2D8 tumor samples) from 28 patients to identify transcriptional signatures (Fig. 1). BOR was determined by RECIST for 23 of the 26 patients with baseline tumor samples: four PR, eight SD, and 11 PD. Twelve patients had paired baseline and C2D8 tumor samples (one PR, five SD, six PD). For transcriptional signature analysis, gene expression was expressed as log2(TPM + 0.1), the base 2 logarithmic transformation of the transcript per million of the gene incremented by a negligible offset. Gene set enrichment analyses were performed on Reactome pathways using Data4Cure [19]. TMB and genes/gene sets potentially associated with response to treatment in baseline tissue samples were identified by linear regression analysis. Genes and gene sets potentially up-regulated by treatment with sasanlimab were identified by differential expression analysis of baseline and on-treatment tumor biopsies [20, 21].
2.5 Assessment of PD-L1 Protein Expression by Immunohistochemistry (IHC)
PD-L1 expression was evaluated in 45 tumor biopsies (32 baseline and 13 C2D8 tumor samples) from 34 patients by IHC (Fig. 1) using the Ventana SP263 assay (Ventana Medical Systems, Oro Valley, AZ, USA) and pathologist scoring. Among the 32 patients with baseline tumor samples, BOR was determined by RECIST for 29 of them: five PR, 12 SD, and 12 PD. Baseline tumor samples from 22 patients were evaluated by both PD-L1 IHC and RNA-Seq. The scoring method allowed determination of PD-L1 expression in tumor cells and immune cells. Results were expressed for tumor cells as percentage of viable tumor cells expressing PD-L1 with discernible membrane staining of any intensity (cytoplasmic staining was not included) and for immune cells as percentage of immune cells with PD-L1 expression/total number of immune cells evaluated.
2.6 Assessment of CD8 Protein Expression by Immunohistochemistry (IHC)
CD8 expression was evaluated by IHC in 48 tumor biopsies (33 baseline and 15 C2D8 tumor samples) from 35 patients (Fig. 1). BOR was determined by RECIST for 30 of the 33 patients with baseline tumor samples: five PR, 12 SD, and 13 PD. IHC sample slides were double labeled for CD8 and panCK (cytokeratin) expression by immunostaining (HistoGenX, Naperville, IL, USA) and assessed using Visiopharm image analysis. The tissue area stained with panCK was analyzed to generate a carcinoma “mask” denoting the tumor region. The cell number and relative surface areas (%) of CD8 tumor-infiltrating T cells within the carcinoma “mask” were then measured in the stromal and in the epithelioid (carcinoma cell nests) tumor compartments using image analysis applications. Analyses were performed for both the invasive margin and the center of the tumor samples.
2.7 Statistical Analyses
Best tumor percentage change from baseline, by RECIST v1.1 [17], was used as response for the biomarker analyses. Association with response of TMB, PD-L1 IHC expression, CD8 IHC expression, and gene set of interest were assessed by linear regression modeling. Due to limitations in sample size, response variables were not pre-adjusted by covariates such as dose level and tumor types.
Individual genes associated with response were identified by the R limma package, using the voom function for gene expression normalization [20]. Gene set expression was defined as the mean expression of the involved single genes. Pathways associated with response were identified by gene ranking-based gene set enrichment analysis (pre-ranked GSEA analysis by Data4Cure on Reactome pathways) of genes ordered by statistics from the association analysis [19].
Differentially expressed genes after treatment were identified by paired analysis on baseline and C2D8 samples using the R limma package [20]. Differentially expressed pathways were identified by pre-ranked GSEA analysis by Data4Cure on Reactome pathways of genes ordered by statistics from the differential analysis.
3 Results
Twenty-three patients received sasanlimab IV at 1 mg/kg (n = 8), 3 mg/kg (n = 8), or 10 mg/kg (n = 7) q3w in the dose-escalation (n = 12) and pharmacodynamic (n = 11) cohorts. In addition, 15 patients received sasanlimab SC 300 mg q4w. Further, two patients treated in dose escalation at the lowest dose level (0.5 mg/kg) were not included in these analyses.
The patients had a diagnosis of advanced disease across a broad range of solid tumors, including adenocarcinoma of the salivary glands (n = 1), endometrial adenocarcinoma (n = 1), malignant melanoma (n = 1), malignant peritoneal neoplasm (n = 2), esophageal adenocarcinoma (n = 2), NSCLC (n = 1), ovarian cancer (n = 15), renal cell carcinoma (n = 1), sarcoma (n = 6), SCLC (n = 3), and SCCHN (n = 7).
BOR was determined by RECIST for 36 of the 40 patients: one patient achieved a complete response (CR), seven had a PR, 13 had SD, and 15 experienced disease progression. The disease control rate (CR + PR + SD) and the PR rate were 60 and 40%, respectively, in evaluable patients with high tumor PD-L1 expression by IHC (≥ 1%, n = 10) versus 52.4 and 4.8%, respectively, in evaluable patients with low tumor PD-L1 IHC expression (< 1%, n = 21) at baseline (PD-L1 expression levels were not known for the patient with CR).
3.1 Baseline Tumor Mutational Burden (TMB)
Assessment of a potential relationship between baseline TMB and best change in tumor burden, by analysis of TMB in available tumor biopsies, showed that a higher TMB at baseline was significantly associated with response to sasanlimab administered IV or SC, across different tumor types, dose levels, and routes of administration (n = 24; p = 0.0134) (Fig. 2).
Baseline TMB and response to treatment with sasanlimab (p = 0.0134). The statistics were calculated by linear regression of TMB on best percentage change from baseline in tumors. The blue solid line represents the regression model fit along with its 95% confidence intervals (dark gray shaded area). Dashed lines indicate the cutoff between PR and SD (30% tumor shrinkage) and the cutoff between SD and disease progression (20% tumor growth). Adj adjusted, PR partial response, SD stable disease, TMB tumor mutational burden
3.2 PD-L1 Protein Expression by Immunohistochemistry (IHC) and PD-L1 RNA Expression in Baseline Tissues
PD-L1 expression was assessed in baseline tumor biopsies by immunohistochemical analysis of SP263 membrane staining in viable tumor cells and in immune cells (n = 29) (Fig. 3A, B, respectively) and by RNA-Seq in non-separated, bulk RNA samples (n = 23) (Fig. 3C).
Analysis of baseline PD-L1 (a–c) and CD8 (d, e) protein/RNA expression and response to treatment with sasanlimab. The statistics were calculated from linear regression of protein/RNA expression on best percentage change from baseline in IHC and RNA-Seq samples. Round dots: baseline tissues with both IHC and RNA-Seq profiling; triangles: baseline tissues with IHC only or RNA-Seq only. PD-L1 protein expression results presented as a percentage viable tumor cells expressing PD-L1 (membrane staining) (p = 0.805) and b percentage immune cells with PD-L1 expression/total immune cells (p = 0.258). c, e RNA-Seq analyses performed in non-separated, bulk RNA samples (p = 0.0294 and p = 0.0343, respectively). d CD8 protein expression evaluated by number of immune cells within the central tumor area (p = 0.79). The blue solid line represents the regression model fit along with its 95% confidence intervals (dark gray shaded area). Adj adjusted, IHC immunohistochemistry, PD-L1 programmed cell death ligand 1, RNA-Seq ribonucleic acid sequencing, TPM transcripts per million
Change in tumor burden following treatment with sasanlimab was significantly associated with PD-L1 RNA levels (p = 0.0294) but not with PD-L1 protein levels detected by IHC in tumor cells and stromal immune cells (p = 0.258–0.805) in this heterogeneous tumor population. Similarly, when evaluating the subset of patients with both IHC and RNA samples available (n = 22), change in tumor burden with sasanlimab showed a stronger association with PD-L1 RNA levels (p = 0.0892) than with PD-L1 protein detected by IHC (p = 0.811–0.926). PD-L1 expression detected by IHC in stromal immune cells correlated to a limited extent with PD-L1 gene expression assessed by RNA-Seq (p = 0.048).
3.3 CD8 Protein Expression by Immunohistochemistry (IHC) and CD8 RNA Expression in Baseline Tissues
CD8 expression was assessed by IHC with multiple endpoints, including immune cell number or area within both the stromal and the intraepithelial compartments of the central tumor and invasive margin areas. Although some trends were observed of increased baseline CD8 levels related to greater tumor shrinkage relative to baseline, none of these parameters showed a significant association. Fig. 3D shows immune cells within the central tumor area as a representative example. However, similar to the results observed for PD-L1 RNA versus protein expression, bulk CD8A gene expression by RNA-Seq was significantly associated with response to treatment (p = 0.0343) across tumor types (Fig. 3E). The rank correlation between CD8 IHC and CD8A RNA expression was 0.89 (p < 0.0001). As only a portion of the samples (n = 17) had both IHC and RNA results, the assessment of the correlation between IHC and RNA may not be robust given the small sample size.
3.4 Differential Expression Analysis of Baseline Biopsies
Genes associated with response to sasanlimab were identified in baseline biopsies (n = 23) from association analysis between gene expression and best percentage change from baseline in tumors (Fig. 4). CTLA4 (encoding cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4)), an immune checkpoint, was among the genes most significantly associated with response to treatment with sasanlimab (Fig. 4B). IFNG (encoding interferon-gamma (IFN-γ)) and the IFN-γ signature gene set (including the 18 genes CD274, CXCR6, TIGIT, CD27, PDCD1LG2, LAG3, NKG7, PSMB10, CMKLR1, CD8A, IDO1, CCL5, CXCL9, HLA-DQA1, CD276, HLA-DRB1, STAT1, and HLA-E) [22] were also significantly associated with better outcome (Fig. 4C, D).
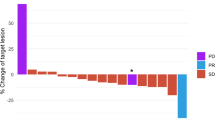
Analysis of genes associated with response to treatment with sasanlimab in baseline biopsies. a Genes positively and negatively associated with improved response (p < 0.05) are shown in green and red, respectively. b, c Dot plots display the association between CTLA4 or IFNG baseline gene expression and best percentage change from baseline in tumors (p < 0.0001 and p = 0.00271, respectively). p values for CTLA and IFNG were generated by limma [20]. d Dot plot of IFN-γ signature expression and best percentage change from baseline in tumors, analyzed by linear regression (p = 0.0201). The blue solid line represents the regression model fit along with its 95% confidence intervals (dark gray shaded area). IFN-γ signature expression was defined as the mean expression of 18 IFN-γ related genes (CD274, CXCR6, TIGIT, CD27, PDCD1LG2, LAG3, NKG7, PSMB10, CMKLR1, CD8A, IDO1, CCL5, CXCL9, HLA-DQA1, CD276, HLA-DRB1, STAT1, HLA-E). Adj adjusted, IFN-γ interferon-gamma, TPM transcripts per million
Gene set enrichment analysis indicated that genes positively associated with response to sasanlimab were enriched in the Reactome [19] IFN-γ and PD-1 signaling pathway gene sets, as well as the cell cycle gene set (Fig. 5) (i.e., genes encoding proteins involved in G1-G1/S mitotic phase, S phase, M phase, and cell cycle checkpoints). Conversely, enrichment in extracellular matrix organization-related pathway genes appeared to be associated with worse outcome (Fig. 5B).
Gene set enrichment analysis by pre-ranked GSEA on results from association analysis between baseline gene expression and response to treatment (Data4Cure with Reactome pathways). a Dots indicate Reactome pathways curated by Data4Cure and lines indicate the relation of the pathways. Network view’s colors indicate the significance of gene set enrichment: burgundy = pathways enriched in genes positively associated with response (good prognosis), green = pathways enriched in genes negatively associated with response (poor prognosis). b Most significantly enriched pathways; bar length shows the statistic from pre-ranked GSEA analysis, where larger absolute values indicate more significant enrichment. Dashed lines indicate q value = 0.05 [18]. APC/C anaphase-promoting complex/cyclosome, Cdc cell-division cycle protein, DNA deoxyribonucleic acid, ECM extracellular matrix, G1 gap 1, GSEA gene set enrichment analysis, HDAC histone deacetylase, M mitosis, PD-1 programmed cell death 1, S synthesis
3.5 Intravenous (IV) and Subcutaneous (SC) Administration of Sasanlimab
When patients were divided by route of administration, no differences were observed between the IV and SC route in terms of association with response to sasanlimab for the biomarkers of interest, including TMB, PD-L1 expression, CD8 expression, and IFN-γ signature score (Online Supplementary Material Resources 1–4).
3.6 Multivariate Analysis
To assess whether baseline TMB and immune gene expression were independent or correlated predictors of response to treatment, 21 samples with both TMB and immune gene expression data available for analysis were evaluated. Higher baseline TMB only, IFNG gene expression only, and IFN-γ gene signature score only were significantly associated with best change in tumor burden (p = 0.0077, p = 0.0116, and p = 0.029, respectively). When baseline TMB and IFNG gene expression were combined as predictors (multivariate model), their effects remained significant, indicating that they were independently associated with change in tumor size. In the multivariate model for TMB and IFNG, the effect was − 8.84 (p = 0.0076) for TMB and − 8.11 (p = 0.0096) for IFNG. In the multivariate model for TMB and IFN-γ gene signature score, the effect was − 8.81 (p = 0.0155) for TMB and − 12.76 (p = 0.0564) for the IFN-γ gene signature. The correlation coefficient was 0.18 between TMB and IFNG gene expression and 0.23 between TMB and IFN-γ gene signature score, indicating a lack of correlation between these markers as predictors of response. Although limited by the small sample size, similar findings were obtained in the analysis of ovarian cancer samples only and of the multiple tumor types on study except for ovarian cancer.
3.7 Pharmacodynamic Effects: Genes Modulated by Treatment with Sasanlimab
Differential expression analysis of paired baseline and on-treatment (C2D8) tumor biopsies (n = 12) showed increased expression of genes enriched in adaptive immune response pathways, including cytokines, chemokines, and chemokine receptors, and decreased expression of genes related to the cell cycle (Figs. 6, 7).
a Genes differentially expressed in on-treatment (C2D8) versus baseline biopsies. Up-regulated and down-regulated genes with p < 0.05 are shown in red and green, respectively. b Dot plots display expression of selected genes in on-treatment versus baseline biopsies: CXCL9 (p = 0.00168); PDCD1 (p = 0.00759); IFNG (p = 0.00727); CD274 (p = 0.211); CTLA4 (p = 0.421); CDKN1B (p = 0.0101). p values were generated by limma [20]. C2D8 Cycle 2, Day 8, CD274 cluster of differentiation 274, CDKN1B cyclin dependent kinase inhibitor 1B, CTLA4 cytotoxic T-lymphocyte-associated protein 4, CXCL9 C-X-C motif chemokine ligand 9, IFNG interferon gamma, PDCD1 programmed cell death protein 1, TPM transcripts per million
Gene set enrichment analysis by pre-ranked GSEA on results from differential analysis between on-treatment and baseline biopsies (Data4Cure with Reactome pathways). a Network view of pathway expression analysis results using statistics from paired differential expression analysis. Significance of pathway enrichment: burgundy = pathways enriched in genes with increased expression after treatment, green = pathways enriched in genes with decreased expression after treatment. b Most significantly enriched pathways. Dashed lines indicate q value = 0.05 [18]. APOBEC3G apolipoprotein B mRNA editing enzyme-catalytic polypeptide-like 3G, AUF1 AU-rich element RNA-binding protein 1, Cdc cell-division cycle protein, CDK cyclin-dependent kinase, CENPA centromere protein A, COP1 constitutive photomorphogenic 1, DNA deoxyribonucleic acid, DVL disheveled proteins, ER endoplasmic reticulum, ERAD endoplasmic-reticulum-associated protein degradation, G1 gap 1, GSEA gene set enrichment analysis, Hh hedgehog, hnRNP heterogenous nuclear ribonucleoprotein, mRNA messenger ribobucleic acid, NF-κB nuclear factor kappa B, PAK p21-activated protein kinase, PD-1 programmed cell death 1, RNP ribonucleoprotein, S synthesis
Gene expression of CXCL9 (a T-cell chemoattractant), PDCD1 (encoding the PD-1 receptor), and IFNG was increased after treatment with sasanlimab, whereas expression of CDKN1B (encoding cyclin-dependent kinase inhibitor 1B, which blocks the cell cycle in the G0/G1 phase) was decreased. No significant changes were observed in CD274 (encoding PD-L1) and CTLA4 gene expression (Fig. 6B).
4 Discussion
The results from this study indicate that tumor responses to the anti-PD-1 monoclonal antibody sasanlimab were associated with baseline TMB, PD-L1, CD8, and IFN-γ gene signature levels, across the multiple dose levels, routes of administration, and broad range of tumor types evaluated in a cohort of 38 patients. In addition, baseline TMB appears to be an independent predictor of response from the other immune-related genes/signatures.
The results on TMB are consistent with prior reports indicating an association between higher TMB at baseline and response to treatment with other immune checkpoint inhibitors (i.e., nivolumab, pembrolizumab) across cancer histologies [23,24,25,26,27,28]. No prior study has demonstrated this correlation with SC administration. A higher mutational burden may generate neoantigenic determinants within tumors and facilitate targeting of the antitumor immune responses restored by PD-1/PD-L1 therapies [23]. Results from a large analysis have shown that patients with the highest somatic TMB (upper 20% in each tumor type) had better survival following treatment with immune checkpoint inhibitors compared with patients with low TMB [24]. However, this TMB cutoff varied among the different histologies analyzed, preventing the identification of a single cutoff value for TMB that might be suitable for routine clinical use [24]. While the small sample size for tumor types in this study may limit generalizations to other cohorts with varying tissue compositions, and the heterogeneity of tumor types may risk the introduction of confounding variables, the data suggest that TMB may be used as a biomarker of response to anti-PD-1 therapy across tumor types [28] and, consistent with these findings, the US Food and Drug Administration has approved pembrolizumab for the treatment of adult and pediatric patients with unresectable or metastatic TMB-high (≥ 10 mutations/megabase) solid tumors, based on the results of the KEYNOTE-158 trial [27].
Currently, PD-L1 levels, determined by IHC analysis, are the key biomarker used to identify patients likely to respond to treatment with anti-PD-1/PD-L1 monoclonal antibodies in some tumor types such as NSCLC and SCCHN, although in other tumor types, such as melanoma, it is of limited use. In addition, clinical benefit has been reported in patients with low or no apparent PD-L1 expression [2, 29,30,31,32]. This raised concerns on the reliability and predictive value of PD-L1 assessments based on available detection methods and the risk of excluding patients from potentially beneficial therapies [30,31,32]. Although PD-L1 levels by a categorical IHC cutoff did appear to enrich for responders in the population with tumor cell PD-L1 expression ≥ 1%, in the analysis, PD-L1 RNA levels measured by RNA-Seq in bulk RNA samples at baseline appeared to be better associated with the magnitude of changes in tumor burden following treatment with sasanlimab than PD-L1 protein expression evaluated by IHC staining with SP263. It is possible that cell surface expression of PD-L1 but not PD-L1 RNA expression levels is influenced by the tissue of origin and it is therefore heterogeneous among tumor types, thus suggesting that PD-L1 RNA is a better biomarker than cell surface protein levels for correlation studies across tumor types. Also, given the sensitivity of RNA-Seq to detect transcript versus IHC to detect protein, there is a much greater dynamic range with RNA-Seq than IHC, particularly at low levels of PD-L1 expression, that can be used for correlation studies.
Expression analysis of genes potentially correlated with response to treatment by pre-ranked GSEA showed that in addition to CD8, higher baseline expression levels of genes related to the IFN-γ and the PD-1 signaling pathways, as well as the cell cycle, were associated with response to sasanlimab across tumor types. Thus, these findings are in agreement with the analysis of gene expression profiles in baseline samples from patients with metastatic melanoma, SCCHN, gastric cancer, and other tumor types, which showed that levels of IFN-γ-responsive genes related to antigen presentation (i.e., CIITA, HLADRA), cytokines/chemokines involved in initiation of inflammation (i.e., CXCR6, CXCL9, CCL5, CCR5), and mediators of cytotoxic activity (i.e., granzymes) predicted response to treatment with pembrolizumab [22]. Furthermore, pre-treatment gene expression signatures tracking IFN-γ activation and T-cell activation were found to be associated with disease control (response or stable disease for ≥ 3 months) in patients with advanced NSCLC, SCCHN, and melanoma, independently of the tumor type and the anti-PD-1 agent administered [33].
In this study, evaluation of pharmacodynamic changes showed increased expression of genes enriched in adaptive immune response pathways, such as cytokines (i.e., IFNG), chemokines (i.e., the T-cell chemoattractant CXCL9), and chemokine receptors, in biopsy tissues from various solid tumor types, following treatment with sasanlimab. Conversely, expression of the cell cycle-associated gene CDKN1B was decreased. Consistent with these findings, adaptive immune gene signatures expressed in early, on-treatment tumor biopsies from patients with metastatic melanoma were previously shown to be associated with response to PD-1 therapy, after prior treatment with a CTLA-4 inhibitor [34]. Most of the genes analyzed and differentially expressed in patients with a clinical response versus non-responders were up-regulated following immune checkpoint inhibition, including IFN-γ pathway effectors, major histocompatibility complex human leukocyte antigen molecules, cytolytic markers, chemokines, and adhesion molecules [34]. Dendritic cell chemoattractant chemokines (i.e., C–C and C–X–C motif chemokine ligands) were also found to contribute to prediction of positive response to anti-PD-1 therapy in a tumor cell genomic analysis conducted in patients with NSCLC treated with pembrolizumab [35].
5 Conclusions
In conclusion, the increased expression of adaptive immune activation genes/pathways in on-treatment biopsies suggests an active, immunomodulatory mechanism for sasanlimab therapy across different tumor types and routes of administration. Further research is required to verify these findings.
References
Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2018;175:313–26.
Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer. 2018;6:8.
Ugurel S, Röhmel J, Ascierto PA, Flaherty KT, Grob JJ, Hauschild A, et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur J Cancer. 2017;83:247–57.
Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, et al. Immunotherapy in non-small cell lung cancer: facts and hopes. Clin Cancer Res. 2019;25:4592–602.
Marur S, Singh H, Mishra-Kalyani P, Larkins E, Keegan P, Sridhara R, et al. FDA analyses of survival in older adults with metastatic non-small cell lung cancer in controlled trials of PD-1/PD-L1 blocking antibodies. Semin Oncol. 2018;45:220–5.
Lalani AA, McGregor BA, Albiges L, Choueiri TK, Motzer R, Powles T, et al. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions. Eur Urol. 2019;75:100–10.
Ghanizada M, Jakobsen KK, Grønhøj C, von Buchwald C. The effects of checkpoint inhibition on head and neck squamous cell carcinoma: a systematic review. Oral Oncol. 2019;90:67–73.
Xu W, Atkins MB, McDermott DF. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol. 2020;17:137–50.
Remon J, Passiglia F, Ahn MJ, Barlesi F, Forde PM, Garon EB, et al. Immune checkpoint inhibitors in thoracic malignancies: review of the existing evidence by an IASLC expert panel and recommendations. J Thorac Oncol. 2020;15:914–47.
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29.
Nowicki TS, Hu-Lieskovan S, Ribas A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J. 2018;24:47–53.
Schoenfeld AJ, Hellmann MD. Acquired resistance to immune checkpoint inhibitors. Cancer Cell. 2020;37:443–55.
Youssef S, Abdiche Y, Nguyen H, Chou J, Chin SM, Kamperschroer C, et al. In vitro properties and pre-clinical activity of PF-06801591, a high-affinity engineered anti-human PD-1. Cancer Res. 2017;77:2667.
Hu-Lieskovan S, Johnson M, Braiteh F, Grilley-Olson J, Chou J, Davda J, et al. Safety, efficacy, pharmacokinetics (PK) and pharmacodynamics (PD) of PF 06801591, an anti-PD1 antibody administered intravenously (IV) or subcutaneously (SC). Ann Oncol. 2017;28:v419.
Johnson ML, Braiteh F, Grilley-Olson JE, Chou J, Davda J, Forgie A, et al. Assessment of subcutaneous vs intravenous administration of anti-PD-1 antibody PF-06801591 in patients with advanced solid tumors: a phase 1 dose-escalation trial. JAMA Oncol. 2019;5:999–1007.
Ji Y, Liu P, Li Y, Bekele BN. A modified toxicity probability interval method for dose-finding trials. Clin Trials. 2010;7:653–63.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
Patwardhan A, Harris J, Leng N, Bartha G, Church DM, Luo S, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med. 2015;7:71.
Biomedical Intelligence Cloud. Data4Cure, Inc., La Jolla, CA, USA. 2020. https://www.data4cure.com. Accessed 12 Oct 2020.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Law CW, Chen Y, Shi W, Smyth GK. Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–40.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8.
Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6.
Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37:992–1000.
Hu-Lieskovan S, Lisberg A, Zaretsky JM, Grogan TR, Rizvi H, Wells DK, et al. Tumor characteristics associated with benefit from pembrolizumab in advanced non-small cell lung cancer. Clin Cancer Res. 2019;25:5061–8.
Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65.
Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593.
Ancevski Hunter K, Socinski MA, Villaruz LC. PD-L1 testing in guiding patient selection for PD-1/PD-L1 inhibitor therapy in lung cancer. Mol Diagn Ther. 2018;22:1–10.
Weinberg BA, Xiu J, Hwang JJ, Shields AF, Salem ME, Marshall JL. Immuno-oncology biomarkers for gastric and gastroesophageal junction adenocarcinoma: why PD-L1 testing may not be enough. Oncologist. 2018;23:1171–7.
Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7:278.
Powles T, Walker J, Williams JA, Bellmunt J. The evolving role of PD-L1 testing in patients with metastatic urothelial carcinoma. Cancer Treat Rev. 2020;82:101925.
Prat A, Navarro A, Paré L, Reguart N, Galván P, Pascual T, et al. Immune-related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res. 2017;77:3540–50.
Chen P-L, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–37.
Brogden KA, Parashar D, Hallier AR, Braun T, Qian F, Rizvi NA, et al. Genomics of NSCLC patients both affirm PD-L1 expression and predict their clinical responses to anti-PD-1 immunotherapy. BMC Cancer. 2018;18:225.
Acknowledgements
The authors thank the patients and their families/caregivers, and the investigators, research nurses, study coordinators, and operations staff who contributed to this study. Medical writing support was provided by Sara Mariani, MD, PhD, and Simon Stones, of Engage Scientific Solutions, and was funded by Pfizer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was sponsored by Pfizer.
Conflict of interest
S. Hu-Lieskovan disclosed consulting honoraria from Amgen, Genmab, Xencor, and Merck; research funding from Bristol Myers Squibb, Merck, and Vaccinex. F. Braiteh disclosed speaker bureau and advisory board honoraria from Pfizer. J. E. Grilley-Olson disclosed no relevant conflict of interest. X. Wang, A. Forgie, V. Bonato, I. A. Jacobs, and J. Chou were employees of and owned stock in Pfizer Inc. at the time of this study. ML Johnson disclosed institutional research funding from AbbVie, Adaptimmune, Amgen, Apexigen, Arcus Biosciences, Array BioPharma, Artios Pharma, AstraZeneca, ATRECA, BeiGene, BerGenBio, Birdie Pharmaceuticals/Seven and Eight Biopharmaceuticals, Boehringer Ingelheim, Calithera Biosciences, Checkpoint Therapeutics, Corvus, Curis, CytomX, Daiichi-Sankyo, Dracen Pharmaceuticals, Dynavax, EMD Serono, Genentech/Roche, Genmab, Genocea, GlaxoSmithKline, Gritstone Oncology, Harpoon Therapeutics, Hengrui Therapeutics, Immunocore, Incyte, Janssen, Lilly, Loxo Oncology, Lycera, Merck, Mirati Therapeutics, Neovia, Novartis, Pfizer, PMV Pharmaceuticals, Regeneron, Ribon Therapeutics, Sanofi, Shattuck Labs, Silicon Therapeutics, Stemcentrx, Syndax, Takeda, Tarveda Therapeutics, TCR2 Therapeutics, TMUNITY Therapeutics, University of Michigan, and WindMIL Therapeutics; spouse role as contract lobbyist for Astellas and Otsuka Pharmaceuticals; and consulting honoraria (to institution) from AbbVie, Amgen, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Calithera, Celgene, Daiichi Sankyo, Editas Medicine, Eisai, EMD Serono, G1 Therapeutics, Genentech/Roche, GlaxoSmithKline, Gritstone Oncology, Ideaya Biosciences, Incyte, Janssen, Lilly, Loxo Oncology, Merck, Mirati Therapeutics, Novartis, Pfizer, Ribon Therapetics, Sanofi and WindMIL Therapeutics.
Ethics approval
The study was approved by the institutional review board or independent ethics committee of the participating institutions and followed the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice guidelines.
Consent to participate
All patients provided written informed consent.
Consent for publication
Not required.
Availability of data and material
Upon request, and subject to certain criteria, conditions, and exceptions (see https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information), Pfizer will provide access to individual de-identified participant data from Pfizer-sponsored global interventional clinical studies conducted for medicines, vaccines, and medical devices (1) for indications that have been approved in the US and/or EU or (2) in programs that have been terminated (i.e., development for all indications has been discontinued). Pfizer will also consider requests for the protocol, data dictionary, and statistical analysis plan. Data may be requested from Pfizer trials 24 months after study completion. The de-identified participant data will be made available to researchers whose proposals meet the research criteria and other conditions, and for which an exception does not apply, via a secure portal. To gain access, data requestors must enter into a data access agreement with Pfizer.
Code availability
Not applicable.
Author contributions
All authors contributed to data analysis and interpretation for this study as well as manuscript development. All authors read and approved the final manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Hu-Lieskovan, S., Braiteh, F., Grilley-Olson, J.E. et al. Association of Tumor Mutational Burden and Immune Gene Expression with Response to PD-1 Blockade by Sasanlimab Across Tumor Types and Routes of Administration. Targ Oncol 16, 773–787 (2021). https://doi.org/10.1007/s11523-021-00833-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11523-021-00833-2