Abstract
Deep learning provides the healthcare industry with the ability to analyse data at exceptional speeds without compromising on accuracy. These techniques are applicable to healthcare domain for accurate and timely prediction. Convolutional neural network is a class of deep learning methods which has become dominant in various computer vision tasks and is attracting interest across a variety of domains, including radiology. Lung diseases such as tuberculosis (TB), bacterial and viral pneumonias, and COVID-19 are not predicted accurately due to availability of very few samples for either of the lung diseases. The disease could be easily diagnosed using X-ray or CT scan images. But the number of images available for each of the disease is not as equally as other resulting in imbalance nature of input data. Conventional supervised machine learning methods do not achieve higher accuracy when trained using a lesser amount of COVID-19 data samples. Image data augmentation is a technique that can be used to artificially expand the size of a training dataset by creating modified versions of images in the dataset. Data augmentation helped reduce overfitting when training a deep neural network. The SMOTE (Synthetic Minority Oversampling Technique) algorithm is used for the purpose of balancing the classes. The novelty in this research work is to apply combined data augmentation and class balance techniques before classification of tuberculosis, pneumonia, and COVID-19. The classification accuracy obtained with the proposed multi-level classification after training the model is recorded as 97.4% for TB and pneumonia and 88% for bacterial, viral, and COVID-19 classifications. The proposed multi-level classification method produced is ~8 to ~10% improvement in classification accuracy when compared with the existing methods in this area of research. The results reveal the fact that the proposed system is scalable to growing medical data and classifies lung diseases and its sub-types in less time with higher accuracy.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Increasing medical abnormalities has led to life insecurity. Detection and treatment of such abnormalities at an early stage can help save lives. When highly dangerous pathogens like viruses reach the lungs, ground-glass opacity in the chest X-rays is observed due to fibrosis in the lungs. Due to the significant differences between X-ray images of an infected and non-infected person, artificial intelligence techniques can be used to identify the presence and severity of the infection. Authors propose a deep learning classification model using convolutional neural networks (CNN) that can analyse the chest X-rays and diagnose deadly lung diseases like pneumonia and tuberculosis and ultimately identify the victims of COVID-19. Motivation behind this work involves two tasks. (1) Most classification methods tend to perform poorly on minority class samples [1]. This is because they aim to optimize the overall accuracy without considering the relative distribution of each class. Hence, a balanced dataset is important for creating a good prediction model. (2) Conventional supervised machine learning methods do not achieve higher accuracy when trained using lesser amount of COVID-19 data samples [2]. A deep neural network achieves improved accuracy compared to other conventional methods. The objectives of this research work are as follows: (1) to solve the class imbalance problem caused by minimal number of COVID-19 images by oversampling using SMOTE, (2) to increase the efficiency and accuracy of diagnosis using deep learning models to classify the chest X-ray images, (3) to help the physicians make chest X-ray diagnosis more accurately and avoid misdiagnosis of TB and pneumonia using computer-aided diagnosis, and (4) to implement multi-level classification, firstly to classify TB and pneumonia, and secondly to classify COVID-19 and bacterial and viral pneumonias.
The novelty of this research work is to perform multi-stage classification of chest X-ray images, to help the physicians make chest X-ray diagnosis more accurately and avoid misdiagnosis of TB and pneumonia using computer-aided diagnosis.
At the first stage, a binary classification of TB and pneumonia was performed.
As COVID-19 is a type of pneumonia, it is further categorized as COVID-19 and viral and bacterial pneumonias in the second stage.
COVID-19-positive images are very few (minority class) compared to TB and pneumonia. The classification will be biased towards the class which has more number of images (called majority class). To deal with this problem, SMOTE technique was employed to oversample the minority class images and generate more images for COVID-19. This could avoid bias nature of learning algorithm towards the class having more images.
Deep learning architectures were applied to classify the type of chest disease more accurately and efficiently.
2 Related work
Chest X-ray classification using deep learning for automated COVID-19 screening [3]
The authors have discussed a classification model that can analyse the chest X-rays and help in the accurate diagnosis of COVID-19. The methodology classifies the chest X-rays into 4 classes normal, pneumonia, tuberculosis (TB), and COVID-19. Furthermore, the X-rays indicating COVID-19 are classified on severity basis into mild, medium, and severe. The deep learning model used for the classification of pneumonia, TB, and normal is VGG16. For the segregation of normal pneumonia and COVID-19, the DenseNet-161 was used. ResNet-18 worked best for severity classification. Their approach allows mass screening of the people using X-rays as a primary validation for COVID-19 [3–5].
The input chest X-ray image is passed through a VGG-16 model and labeled as normal, tuberculosis, or pneumonia. Furthermore, the pneumonia images are passed through a DenseNet-161 model and categorized as normal pneumonia or COVID-19. The COVID-19 images are passed through a ResNet18 model and classified as severe, medium, or mild [6, 7].
The input size of the image for the VGG-16 model is 64 × 64 × 1. After this, the image passes through a 2D convolution layer with dimensions 224 × 224. Then, it passes through the next layer with dimensions 112 × 112. After this, it passes to consecutive convolution layers along with MaxPooling with dimension changes 56 × 56, 28 × 28, and 14 × 14. Then, it goes to a MaxPooling layer with dimension change 7 × 7. After a series of 2500, 4096, and 4096 fully connected layers, the X-rays are classified into normal X-ray, tuberculosis-affected X-ray, and pneumonia-affected X-Ray [8].
The input size of the image for the DenseNet-161 model is 64 × 64 × 1. After which, the image passes through a 2D convolutional layer with dimensions 112 × 112. Then, it passes through the next layer with dimensions 56 × 56. After passing through the consecutive layers, the dimensions change from 56 × 56 to 28 × 28, 14 × 14, and finally, 7 × 7. After 1000 fully connected layers, the X-rays are classified into normal pneumonia and COVID-19.
The input size of the image for the ResNet-18 model is 64 × 64 × 1. After this, the image passes through a 2D convolution layer with dimensions 112 × 112 × 64. Then, it passes through the next layer with dimensions 56 × 56 × 64. After this, it passes to consecutive 2D convolution layers with dimension changes 28 × 28 × 128, 14 × 14 × 256, and 7 × 7 × 512. After 1000 fully connected layers, the X-rays are classified into severe, medium, and mild [9, 10].
A multi-level classification model was introduced in previous work [3]. They did separate models for separate classification tasks, so that the misclassification of the lung diseases tuberculosis and pneumonia can be eliminated first and pneumonia-classified patients only can be concentrated for further classifications. So, the multi-level classification model was proposed for the classification of lung diseases.
Pneumonia diagnosis using deep learning [11]
The authors have discussed about classification of chest X-ray images to bacterial pneumonia, viral pneumonia, and normal using the VGG pretrained model which has 16 convolution layers followed by a connected hidden layer and dropout layers in between. Dropout layers help to regularize the model to prevent the model from overfitting. A dataset has more than nine thousand chest X-ray images for the classes bacterial pneumonia, viral pneumonia, and normal chest X-rays. This particular model which has CNN layers is able to perform better mainly due to the deep structure of CNN that uses the power of extracting different level features, which resulted in a better generalization capability.
Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans [12]
This study includes eleven pretrained convolutional neural network models, such as Visual Geometry Group (VGG) network and residual neural network (ResNet). They have been successfully tested and evaluated on a public X-ray image dataset for normal and three diseased cases. The results of proposed cascaded classifiers showed that VGG16, ResNet50V2, and dense neural network (DenseNet169) models achieved the best detection accuracy of COVID-19, viral (non-COVID-19) pneumonia, and bacterial pneumonia images, respectively [12].
Viral and bacterial pneumonia detection using artificial intelligence in the era of COVID-19 [13]
The authors have discussed a classification model to classify COVID-19, non-COVID, bacterial pneumonia, viral pneumonia, and normal. The paper proposed that pretrained AlexNet model is employed due to its high accuracy in carrying out feature extraction and image classification. With regard to the classification of COVID-19 and normal CXR images, it was observed that the model provided significantly better performance compared to studies that utilized small amounts of dataset such as Mahmud et al. (2020) and models developed from scratch.
A deep learning approach to detect COVID-19 coronavirus with X-ray images [14]
The authors have discussed about classifying chest X-ray images into viral pneumonia, bacterial pneumonia, and normal cases using ResNet50 deep network architecture at the first stage, and, furthermore, viral pneumonia samples were classified into COVID-19 induced pneumonia and other viral pneumonias with ResNet101 architecture. The limited number of COVID-19 images makes it a bit difficult to train the deep learning models from scratch, which is overcome by using deep transfer learning methods in the current study.
3 Proposed multi-level classification model for classification of lung diseases
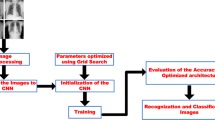
The proposed system has two models (multi-level classification): first model for classifying tuberculosis (TB) and pneumonia and the second model for classifying the types of pneumonia such as bacterial pneumonia, viral pneumonia, and COVID-19 from taking the pneumonia-classified X-ray images (output of the first model) as depicted in Fig. 1.
Since the dataset for the TB, pneumonia, and pneumonia types (first, second levels) are less when compared with datasets for other classification tasks, to improve the classification performance of the model, data augmentation and oversampling techniques are applied to the training set. In the dataset considered under current work [15–17], the data is biased for the pneumonia class; to solve this bias problem, authors have compared with class balancing techniques and have found that SMOTE performs well for class balancing, so authors have used SMOTE to overcome the problem from our dataset.
For the accurate classification of lung diseases, authors have proposed this method of multi-level classification.
-
In first level classification — classification of images into pneumonia and tuberculosis.
-
In second level classification — further classification into viral and bacterial pneumonias and COVID-19.
This is the novel idea authors have proposed in this model.
Data augmentation techniques such as image shifts — width_shift_range and height_shift_range; image flips — horizontal_flip and vertical_flip; image rotations — rotation_range; and image zoom — zoom_range are done for the training set for both the models [18, 19]. The data augmentation and data splitting are depicted in Fig. 2.
Since the dataset is unbalanced, some classes are having a smaller number of images compared to other classes. For balancing the dataset, oversampling techniques such as SMOTE and Borderline-SMOTE SVM (SVM SMOTE) are used for increasing the number of samples in the minority classes [20–24].
After balancing the dataset, a convolutional neural network model was built and trained for the pneumonia and tuberculosis datasets. The model was tested against a test set. The images which are classified as “pneumonia” were saved and are to be used as a test set for the second model.
Another convolutional neural network was built and trained using the bacterial pneumonia, viral pneumonia, and COVID-19 for the second-level classification. And this model was tested by the pneumonia-classified images (from the first model). The proposed system is shown in Fig. 3.
3.1 SMOTE
Synthetic Minority Oversampling Technique (SMOTE) is an oversampling approach in which the minority class is oversampled by creating “synthetic” examples rather than by oversampling with replacement [25, 26].
Algorithm
-
1.
Setting the minority class set A, for each x in A, the k-nearest neighbours of x are obtained by calculating the Euclidean distance between x and every other sample in set A.
-
2.
The sampling rate N is set according to the imbalanced proportion. For each x in A, N examples (i.e., x1, x2, … xn) are randomly selected from its k-nearest neighbours and construct the set A1.
-
3.
For each example xk in A1 (k = 1, 2, 3 … N), the following formula is used to generate a new example: ×′ = x + rand(0,1) × | x − xK | in which rand(0,1) represents the random number between 0 and 1.
Often, real-world datasets are predominantly composed of “normal” examples with only a small percentage of “abnormal” or “interesting” examples. With SMOTE, one can determine the ratio of the oversampling or specifies the class to be oversampled. We can also control the randomization of the algorithm by specifying it in the function’s argument. Other optional parameters like number of nearest neighbours can be specified. SMOTE synthesizes new minority instances between existing minority instances.
SMOTE algorithm is performed here which oversamples the specified class and produces synthetic samples. Sampling strategy is specified which determines the ratio of the oversampling or specifies the class to be oversampled. The randomization of the algorithm can be controlled by specifying it in the function’s argument.
The undersampling will reduce the number of images which would affect the model accuracy.
Bootstrapping would select samples in random which would result in probability of selecting the samples from the class where many images will be high compared to selecting images from class with very few images.
It is necessary to deal with the class which has very few images and remove the bias towards classes during training the model.
Other optional parameters like number of nearest neighbours can be specified. SMOTE synthesizes new minority instances between existing minority instances [27]. It generates the virtual training records by linear interpolation for the minority class. Data balancing using SMOTE is depicted in Fig. 4. Table 1 shows the class distribution of images for tuberculosis and pneumonia.
3.2 Borderline-SMOTE SVM or SVM SMOTE
Borderline-SMOTE SVM is a variant of SMOTE. In this SMOTE type, Borderline-SMOTE, support vector machine (SVM) algorithm is used instead of a KNN to identify misclassified examples on the decision boundary. An SVM is used to locate the decision boundary defined by the support vectors and examples in the minority class that close to the support vectors becomes the focus for generating synthetic examples.
The main difference between SVM-SMOTE and the other SMOTE is that instead of using K-nearest neighbours to identify the misclassification in the Borderline-SMOTE, the technique would incorporate the SVM algorithm [21].
In the SVM-SMOTE, the borderline area is approximated by the support vectors after training SVMs classifier on the original training set. Synthetic data will be randomly created along the lines joining each minority class support vector with a number of its nearest neighbours [22].
In addition to using an SVM, the technique attempts to select regions where there are fewer examples of the minority class and tries to extrapolate towards the class boundary. This variation can be implemented via the SVM SMOTE class from the imbalanced-learn library. Table 2 shows the class distribution of types of pneumonia.
3.3 Construction of DNN model
A convolutional deep neural network model is built in which we have used 1 input layer, 1 output layer with Softmax activation layer, and 5 hidden layers adding totally to 7 layers.
The hidden layers are:
-
Conv2D layer with ReLu activation layer
-
MaxPooling layer
-
Flatten layer
-
Dense layer (300 neurons) with ReLu activation layer
-
Dense layer (100 neurons) with ReLu activation layer
Table 3 shows how the number of neurons employed in the hidden layers of the DNN impacts the accuracy of the second classification model (accuracy of COVID-19 diagnosis).
While having the number of layers as 400, the model performs well rather than increasing or decreasing from 400. So the number of neurons used in the hidden layers was 400.
The number of neurons used in the input layer and the output layer and the number of hidden layers deployed in different datasets are tabulated correspondingly in Table 4.
4 Materials
The datasets, namely the TB and pneumonia dataset, and the types of pneumonia dataset are used for the implementation. In the class distribution, the number of features in each dataset is tabulated in Table 5. From Table 5, it can be seen that the first dataset has a moderate class imbalance and the second dataset has a high class imbalance. The number of sample and class distribution of images is tabulated in Table 5. The chest X-ray images were extracted from Kaggle database, chest X-ray images (pneumonia) [1], tuberculosis (TB) chest X-ray database [28], and curated chest X-ray image dataset for COVID-19 [2].
5 Results
-
(i)
Tuberculosis and pneumonia classification
The dataset was split into 70% for training and 30% for testing. After applying image augmentation and oversampling techniques (SMOTE), a convolutional neural network was built and trained for this dataset and achieved 97.4% accuracy.
-
(ii)
Bacterial pneumonia, viral pneumonia, and COVID-19 classification
The dataset was split into 70% for training and 30% for testing. After applying image augmentation and oversampling techniques (SMOTE), a convolutional neural network was built and trained for this dataset and achieved 88% accuracy. The hyperparameter tuning is mentioned as follows.
The number of nodes in each layer is determined by the experimental basis by adjusting the number in order to reduce the misclassification rate.
Initial learning rate: 0.1
Optimizers used are as follows:
At the hidden layers: ReLu and Softmax
At the output layer: Adam
Loss function: Categorical cross-entropy
Number of epochs: 50 to 200
After applying oversampling techniques, misclassification was reduced and the accuracy was improved for both the classification models. The performance metrics of both levels of classification is tabulated in Tables 6 and 7.
The increase in images will improve accuracy of the model as the model gets trained with more images. But class imbalance will overfit the model to the class which has more number of images. To overcome these bias, SMOTE is applied to balance the number of images in each class so that the bias is removed during classification.
Table 6 shows 3% increase in performance for the first level classification of TB and pneumonia.
Table 7 shows 10 to 15% increase in performance for the second level of classification between viral and bacterial pneumonias and COVID-19.
The TB and pneumonia classification model (first model) was tested against 1646 images. Out of 1646 images, 988 images were classified as “pneumonia”, 658 images were classified as “tuberculosis.” The images that are classified as “pneumonia” from the first model were used as the testing set for the second model (types of pneumonia classification). Out of 988 images, the model classified 541 images as “bacterial pneumonia” and 175 images as “viral pneumonia” and 272 images as “COVID-19.”
Data augmentation and SMOTE techniques have improved the multi-class classification accuracy. Prior to data balancing, the accuracy obtained was 76.3% and post-data balancing the accuracy was 87.1%. This accuracy is with respect to the three-class classification of COVID-19 and bacterial and viral pneumonias. Nevertheless, the binary classification of these three types of chest diseases yielded classification accuracy ~95 to ~98% which was detailed in Section 6 in Table 9.
5.1 Comparison of existing models vs proposed model
The efficiency of existing models was compared with our proposed model’s efficiency in terms of the performance metric accuracy in percentage.
From the above Table 8, it is evident that our proposed model has higher efficiency of 8–11% when compared to the performance of existing models. The framework mentioned in the paper [31] was designed, trained, and validated to identify four classes of CXR images, namely healthy, bacterial pneumonia, viral pneumonia, and COVID-19. The experimental results pose the proposed framework as a potential candidate for COVID-19 disease diagnosis using CXR images, with training, validation, and testing accuracy of 92.4%, 88.24%, and 87.13%, respectively, in four-class classification.
6 Discussions
The other methods to improve classification accuracy are feature selection and autoencoders which are discussed below.
6.1 Feature selection
Irrelevant or partially relevant features can negatively impact model performance. Feature selection is the process where you automatically or manually select those features which contribute most to your prediction variable or output in which you are interested in. Having irrelevant features in your data can decrease the accuracy of the models and make your model learn based on irrelevant features. Some of the advantages of feature selection are,
-
Reduces overfitting: Less redundant data means less opportunity to make decisions based on noise.
-
Improves accuracy: Less misleading data means modeling accuracy improves.
-
Reduces training time: Fewer data points reduce algorithm complexity and algorithms train faster.
6.2 Autoencoders
The current research presents a computer-aided detection model utilizing chest X-ray images for combating the pandemic. The research work in [32] uses features extracted from pretrained networks along with sparse autoencoder for dimensionality reduction and a feed forward neural network (FFNN) for the detection of COVID-19. Autoencoders are proposed as a solution for feature selection. DL techniques used in COVID-19 have also been categorized into seven main distinct categories as long short-term memory networks (LSTM), self-organizing maps (SOMs), conventional neural networks (CNNs), generative adversarial networks (GANs), recurrent neural networks (RNNs), autoencoders, and hybrid approaches [33, 34]. The research work in [35] introduces a novel unsupervised DL-based variational autoencoder (UDL-VAE) model for COVID-19 detection and classification. Autoencoders are used for feature selection to uncover existing nonlinear relationships between features. Autoencoders are unsupervised learners that can learn to compress and encode data and then learn how to reconstruct the data back from the reduced encoded representation. The resulting representation is as close as possible to the original input. In other words, we are taking the feature space and reducing its dimensionality and then reconstructing it from its reduced format. The principle of autoencoders helps us by reducing the data dimensions, learning how to ignore the noise in them, and moreover, knowing which features can best help us reconstruct the data. So, after reconstructing the data, first use the weight matrix of the autoencoder that connects the input feature layer to the reduced layer:
-
If a feature contributes little to the representation of others, it means that the corresponding weight squared (w2) is close to zero.
-
On the other hand, if the feature plays an important role in the representation of other features, the corresponding weight must be significant.
One of the main drawbacks of this method arises from the simplicity of the autoencoder, where a simple single-layer autoencoder cannot model complex non-linear feature dependencies. Feature selection was tried before a multi-level classification module for experimental purposes just to compare the results and see if the efficiency (accuracy) of the model gets better. But feature selection did not work well for our model. The accuracy was achieved only about 77% after applying the feature selection step, whereas without feature selection, our model achieved a higher accuracy of 87%.
Furthermore, the binary classification on two sets of disease types was performed and the results obtained are tabulated in Table 9.
As COVID-19 is a type of viral pneumonia, the accurate categorization was bit low as 95% while bacterial pneumonia vs COVID-19 yielded 98% and bacterial vs viral pneumonia yielded 97%.
7 Conclusion
In our proposed model, the class imbalance problem was handled with the help of SMOTE and SVM-SMOTE, an oversampling algorithm to even out the class distribution. SMOTE performance was evaluated on two datasets: (1) TB and pneumonia multi-class dataset which was moderately imbalanced but highly dimensional and (2) types of pneumonia multi-class dataset which was highly imbalanced. The evaluated results after applying SMOTE were compared with results obtained from before and after applying SMOTE using deep neural network as the classifier. The comparison showed that SMOTE performs significantly better using imbalanced data as it is as there was a 10% increase in the accuracy of the model. The algorithm effectively handled the class imbalance problem. The explanation for this improvement in performance relates to the ability of SVM-SMOTE to generate artificial data in safe areas of the input space, while, at the same time, aggressively increasing the diversity of the generated instances. SVM-SMOTE can be a useful tool for researchers and practitioners since it results in the generation of high-quality artificial data and only requires the tuning of a small number of parameters.
Deep learning can be a very useful tool in the medical industry for the detection of diseases just by processing the images of the chest X-rays and feeding it as an input to the model. The chest X-rays are classified into different classes such as pneumonia, tuberculosis, and COVID-19. Mass screening of people for detection of COVID-19 can be done effectively with our proposed model. It will help yield faster and accurate results and will be cost-effective as compared to the conventional RT-PCR method. This approach can be implemented at the local level and in the rural areas where adequate facilities are absent. The limitation of the current work is that the features used for classifying the chest diseases were not investigated. The feature engineering has to be performed for obtained in the biological feature relevance for identifying the type of chest disease.
Future research directions will include the exploration of image data augmentation techniques to improve accuracy even more, while avoiding overfitting. To expand the size of the middle convolutional layers and making the stride and filter size on the first layer smaller. Use GoogLeNet to reduce the number of parameters in the network. Add more data augmentation techniques to increase the efficiency of training. Design an online application to diagnose pneumonia in chest X-ray images, to serve the largest possible number of slums who lack health care and could not afford for the diagnosis.
References
Kumar P, Bhatnagar R, Gaur K, Bhatnagar A (2021) Classification of imbalanced data: review of methods and applications. In IOP conference series: materials science and engineering, vol 1099, No. 1. IOP Publishing, p 012077
Foody GM, McCulloch MB, Yates WB (1995) The effect of training set size and composition on artificial neural network classification. Int J Remote Sens 16(9):1707–1723
Shelke A, Inamdar M, Shah V, Tiwari A, Hussain A, Chafekar T, Mehendale N (2021) Chest X-ray classification using deep learning for automated COVID-19 screening. SN Comput Sci 2(4):1–9
Rahman T, Chowdhury ME, Khandakar A, Islam KR, Islam KF, Mahbub ZB et al (2020) Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl Sci 10(9):3233
Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL et al (2018) Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172(5):1122–1131
Qjidaa M, Mechbal Y, Ben-Fares A, Amakdouf H, Maaroufi M, Alami B, Qjidaa H (2020) Early detection of COVID19 by deep learning transfer model for populations in isolated rural areas. In 2020 International Conference on Intelligent Systems and Computer Vision (ISCV). IEEE, pp 1–5
Rajaraman S, Candemir S, Kim I, Thoma G, Antani S (2018) Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl Sci 8(10):1715
Liam CK, Pang YK, Poosparajah S (2006) Pulmonary tuberculosis presenting as community-acquired pneumonia. Respirology 11(6):786–792
Bar Y, Diamant I, Wolf L, Lieberman S, Konen E, Greenspan H (2018) Chest pathology identification using deep feature selection with non-medical training. Comput Methods Biomech Biomed Engin: Imaging Visualization 6(3):259–263
Sharma S, Sharma S, Athaiya A (2017) Activation functions in neural networks. Towards Data Sci 6(12):310–316
Barhoom, Alaa MA (2019) Pneumonia diagnosis using deep learning, PhD dissertation., Al-Azhar University-Gaza
Karar ME, Hemdan EED, Shouman MA (2021) Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Complex Intell Syst 7(1):235–247
Ozsoz M, Ibrahim AU, Serte S, Al-Turjman F, Yakoi PS (2020) Viral and bacterial pneumonia detection using artificial intelligence in the era of COVID-19
Jain G, Mittal D, Thakur D, Mittal MK (2020) A deep learning approach to detect Covid-19 coronavirus with X-ray images. Biocybern Biomed Eng 40(4):1391–1405
Chest X-ray images (pneumonia). https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. Accessed 12 July 2022
Tuberculosis (TB) chest X-ray database. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. Accessed 12 July 2022.
Curated chest X-ray image dataset for COVID-19. https://www.kaggle.com/unaissait/curated-chest-xray-image-dataset-for-covid19. Accessed 12 July 2022
Perez L, Wang J (2017) The effectiveness of data augmentation in image classification using deep learning. arXiv preprint arXiv:1712.04621
Mikołajczyk A, Grochowski M (2018) Data augmentation for improving deep learning in image classification problem. In 2018 international interdisciplinary PhD workshop (IIPhDW). IEEE, pp 117–122
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
Wang Q, Luo Z, Huang J, Feng Y, Liu Z (2017) A novel ensemble method for imbalanced data learning: bagging of extrapolation-SMOTE SVM. Comput Intell Neurosci 2017
Nguyen HM, Cooper EW, Kamei K (2011) Borderline over-sampling for imbalanced data classification. J Knowl Eng Soft Data Paradig 3(1):4–21
Fernández A, Garcia S, Herrera F, Chawla NV (2018) SMOTE for learning from imbalanced data: progress and challenges, marking the 15-year anniversary. J Artif Intell Res 61:863–905
Jeatrakul P, Wong KW, Fung CC (2010) Classification of imbalanced data by combining the complementary neural network and SMOTE algorithm. International Conference on Neural Information Processing. Springer, Berlin, Heidelberg, pp 152–159
Özdemir A, Polat K, Alhudhaif A (2021) Classification of imbalanced hyperspectral images using SMOTE-based deep learning methods. Expert Syst Appl 178:114986
Li Y, Wang Y, Li T, Li B, Lan X (2021) SP-SMOTE: a novel space partitioning based synthetic minority oversampling technique. Knowl-Based Syst 228:107269
Li J, Zhu Q, Wu Q, Fan Z (2021) A novel oversampling technique for class-imbalanced learning based on SMOTE and natural neighbors. Inf Sci 565:438–455
Waqar M, Dawood H, Dawood H, Majeed N, Banjar A, Alharbey R (2021) An efficient SMOTE-based deep learning model for heart attack prediction. Scientific Programming, 2021
Nagamounika R, Sri CNSV, Harshitha A, Tejaswi KL, Lakshmi PRSM (2020) Prediction of pneumonia disease by using deep convolutional neural networks. J Eng Sci Criterion 17:18
Oh Y, Park S, Ye JC (2020) Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging 39(8):2688–2700
Johri S, Goyal M, Jain S, Baranwal M, Kumar V, Upadhyay R (2021) A novel machine learning-based analytical framework for automatic detection of COVID-19 using chest X-ray images. Int J Imaging Syst Technol 31(3):1105–1119
Gayathri JL, Abraham B, Sujarani MS, Nair MS (2022) A computer-aided diagnosis system for the classification of COVID-19 and non-COVID-19 pneumonia on chest X-ray images by integrating CNN with sparse autoencoder and feed forward neural network. Comput Biol Med 141:105134
Heidari A, Navimipour NJ, Unal M, Toumaj S (2021) The COVID-19 epidemic analysis and diagnosis using deep learning: a systematic literature review and future directions. Comput Biol Med 105141
Chouhan V, Singh SK, Khamparia A, Gupta D, Tiwari P, Moreira C, De Albuquerque VHC (2020) A novel transfer learning based approach for pneumonia detection in chest X-ray images. Applied Sciences 10(2):559
Mansour RF, Escorcia-Gutierrez J, Gamarra M, Gupta D, Castillo O, Kumar S (2021) Unsupervised deep learning based variational autoencoder model for COVID-19 diagnosis and classification. Pattern Recogn Lett 151:267–274
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent is not necessary as this article does not involve human or animal participants.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Venkataramana, L., Prasad, D.V.V., Saraswathi, S. et al. Classification of COVID-19 from tuberculosis and pneumonia using deep learning techniques. Med Biol Eng Comput 60, 2681–2691 (2022). https://doi.org/10.1007/s11517-022-02632-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02632-x