Abstract
A crucial step in the battle against the coronavirus disease 2019 (Covid-19) pandemic is efficient screening of the Covid affected patients. Deep learning models are used to improve the manual judgements made by healthcare professionals in classifying Chest X-Ray (CXR) images into Covid pneumonia, other viral/bacterial pneumonia, and normal images. This work uses two open source CXR image dataset having a total of 15,153 (dataset 1), and 4575 (dataset 2) images respectively. We trained three neural network models with a balanced subset of dataset 1 (1345 images per class), balanced dataset 2 (1525 images per class), and an unbalanced full dataset 1. The models used are VGG16 and Inception Resnet (IR) using transfer learning and a tailor made Convolutional Neural Network (CNN). The first model, VGG16 gives an accuracy, sensitivity, specificity, and F1 score of 96%, 97.8%, 95.92%, 97% respectively. The second model, IR gives an accuracy, sensitivity, specificity and F1 score of 97%, 98.51%, 97.28%, 99% respectively. The third and best proposed model, CNN gives an accuracy, sensitivity, specificity, and F1 score of 97%, 98.21%, 96.62%, 98% respectively. These performance metrics were obtained for the balanced dataset 1 and all models used 80:10:10 cross validation technique. The highest accuracy using CNN for all the three datasets are 97%, 96%, and 93% respectively. Gradient-weighted Class Activation Mapping (Grad-CAM) is used to ensure that the model uses genuine pathology markers to generalize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Covid-19 is an infectious disease caused by Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) virus causing respiratory illness which can be spread from person-to-person. Covid-19 was first identified in Wuhan, China in December 2019 prior to becoming a global health crisis. At the time of writing this article, 238,521,855 Covid confirmed cases have been reported globally as per World Health Organization (WHO) on 13th October 2021 and 4,863,818 deaths.
The positive-sense Ribonucleic Acid (RNA) viruses- Coronaviruses, are named as it goes in Latin, corona = crown because of the crown like spikes on its surface [1]. Based on their genomic structure they have 4 main subsets namely alpha, beta, gamma, and delta. The Wuhan coronavirus called the novel corona virus 2019 were identified as beta coronavirus in humans. According to the Centers for Disease Control and Prevention (CDC), the early outbreak was thought to have a link to a large seafood and live animal market. However, later cases with no link to the market confirmed person- to-person transmission of the disease. Additionally, travel-related exportation of cases occurred. The disease caused by the novel corona virus (also called SARS-CoV-2) is termed as coronavirus disease-19 (Covid-2019). The SARS-CoV-2 is believed to be originated from the bats. Reference [1] gives information about the SARS-CoV-2. Prior to Covid-19, the other corona virus infections associated with human disease were SARS-CoV in 2002 at Guangdong Province, China and the Middle East respiratory syndrome coronavirus (MERS- CoV) in 2013 at Saudi Arabia. All the above three infectious diseases are caused by beta coronavirus [2]. Even though both SARS-CoV-2 and SARS-CoV belong to the beta subgroup, the transmission ability of SARS-CoV-2 is more than SARS-CoV [3]. The SARS-CoV-2 has a lower fatality but spreads faster than its priors: the Middle East Respiratory Syndrome coronavirus (MERS-CoV) and SARS-CoV. Reference [4] also mentions about the corona virus history, origin, spread, epidemiology, pathogenesis, clinical features, diagnosis, treatment, and prevention.
Covid-19 primarily affects the respiratory system. SARS-CoV-2 infection may be asymptomatic or it may cause a wide spectrum of symptoms, such as mild symptoms of upper respiratory tract infection or severe illness leading to acute respiratory failure and death. The use of imaging modalities such as CXR, chest Computed Tomography (CT) remains an essential tool in conjunction with molecular and serological diagnosis of Covid-19. Like other pneumonias, Covid-19 pneumonia causes the density of the lungs to increase. This may be seen as whiteness in the lungs on radiography which, depending on the severity of the pneumonia, obscures the lung markings that are normally seen; however, this may be delayed in appearing or absent [5]. Covid chest radiography reports show multilobar involvement, asymmetric patchy or diffuse airspace opacities and peripheral airspace opacities [6]. Ground glass is the frequently reported, among opacities. The ground glass opacity is seen at the pulmonary vessel edges during the early course of Covid-19. Ground glass opacities can be difficult to observe and not specific to Covid-19 thus requiring radiologist confirmation. As Covid cases increase and with lack of trained radiologists, automated methods can assist in the diagnosis procedure [6].
According to WHO, the common symptoms of Covid-19 are shortness of breath, fever, cold, acute respiratory syndrome, cough, loss of taste, sore throat, muscle pain, headache, fatigue etc. Patient recovery is estimated to be within two weeks for mild Covid case but it may take up to 6 weeks for severe cases. But due to prospective genetic mutation shown by the virus, the recovery time extended. The symptoms vary from mild to moderate respiratory illness but could develop into a serious illness for people with cancer, cardiovascular diseases, chronic respiratory disease and diabetes. It has been advised to practice respiratory etiquettes to prevent escalation of Covid-19 by transmission through the nose discharges or droplets of saliva by cough or sneeze from the infected person.
In order to lower the Covid-19 transmission it is important to detect the virus infected patients early so that they can be quarantined and treated sooner. The most reliable method to date, for detecting Covid is a laboratory-based procedure called the Reverse Transcriptase-Polymerase Chain Rection (RT-PCR) testing. The nasopharyngeal swabs are preferred but their accuracy is 73.3%. It is influenced by disease severity and the onset of symptoms [7].
The SARS-CoV-2 RNA is detected in this test, from the respiratory specimens. Though RT-PCR testing is highly specific, it has very low sensitivity [8, 9]. The sensitivity of RTPCR varied from 30 to 70% whereas CXR and CT scans have sensitivity values 69% and 98% respectively. As per reference [10] the accuracies of Machine Learning (ML) and Deep Learning (DL) models ranged from 76 to 99 percent. New research show that CT has 98% sensitivity for Covid infection compared to 71% in RTPCR [11]. Another drawback of the RTPCR test is that during the early stage of infection, it has low positive rate [12]. In general, the test results of a real time RTPCR process takes 4–6 h approximately. This could be even longer in remote and rural areas. Reference [13] states that RTPR had a turnaround time of 4–48 h also. Added to this time, in scenarios where RT-PCR kits are limited in supply, the delay to identify Covid patients at an early stage increases and this inadvertently results in spread of the infection. To subjugate this delay, alternate screening methods were adopted. Hence chest radiographic examination is an alternate screening method where CXR or CT images are used to find visual features that characterise the SARS-CoV-2 viral infection. CXR and CT images show pathological findings at the early stages of Covid-19. These findings could still overlap with the other viral diseases like MERS, SARS, H1N1 or influenza [14].
When it comes to competition between CXR and CT scanning methods, their pros and cons are as follows: CT is costly and causes more radiation exposure whereas CXR is commonly used and cost effective. Though CT images give more feature details, CXR imaging comes into major play, in situations of lack of resources and in greatly affected areas, amid the global pandemic situation. CXR imaging is easily available even at small clinical and imaging centres. When considering developing countries with highly infectious areas, CXR imaging is more accessible than CT imaging especially because of the higher equipment and maintenance costs of CT. There are portable CXR systems available on the contrary to the fixed CT scanners. Easy accessibility and availability have made CXR the most common imaging modality during the pandemic. This aids the allocation of hospital resources efficiently.
In short, CXR has lower sensitivity compared to CT or RT-PCR in early/mild infection. In addition, findings on CXR are not specific and overlap with other infections. CXR has several advantages over CT such as less ionizing radiation, rapid data acquisition, availability in the Intensive Care Units (ICU), and portability [15].
Initial studies show that a Covid-19 infected patient shows abnormalities i.e. specific radiological patterns in their CXR images [11]. CXR and CT scans require an expert radiologist as well as, enough time to visually examine and decide on crucial features that classify the images into infectious categories or normal. One must also agree to the fact that it becomes difficult for amateur radiologists to detect inconsiderable discrepancy with bare eyes due to overlapping features and similar visual cues of the inflammatory and infectious lung diseases. Manual reading of a CT takes 15 min whereas image analysis by Artificial Intelligence (AI) algorithms takes only a few seconds [16]. Specific manifestations corresponding to Covid are revealed in a CT and CXR scan but since there can be an overlap of Covid with other lung infections, it becomes challenging to diagnose Covid in a human centred manner. In such a scenario a computer aided diagnostic system that can accurately classify CXR images into categories of infectious disease is favourable, to aid the radiologist with intelligent solutions and save time.
Various Covid studies expose the fact that geriatrics and individuals with a past record of medical problems, precisely earlier cases of pneumonia are more prone to developing serious complications when infected by Covid. Similarity between Covid and pneumonia lies in the fact that both diseases target the lungs to spread the infection and a CXR is performed to discern the infected regions in the lungs [17]. The nonCovid-19 case of pneumonia also exhibits a representative appearance like that of Covid-19 [18].
A type of lung infection caused by virus, is the viral pneumonia accounting to 30% of pneumonia cases [19]. It can be healed and is mild whereas the Covid-19 virus causes severe pneumonia. In Wuhan, the virus was causing a series of unknown pneumonia cases initially [20]. The opacity in lung radiography can be due to Pneumonia, lung cancer, volume loss, pulmonary bleeding, edema, hence observing patterns corresponding to pneumonia is a hard task [21]. The differential diagnosis of Covid and pneumonia is challenging for a radiologist due to feature similarity of CXR images. Reference [18] assessed the radiologist’s performance in China and US to concluded that for discriminating Covid-19 from viral pneumonia, radiologists often had moderate sensitivity and high specificity (in chest CT). The proposed hypothesis states that a deep learning classifier can authentically differentiate between CXRs with Covid and pneumonia.
2 Literature survey
The literature survey focussed on CXR images to classify them into 3 classes of Covid, pneumonia and healthy. When listing two class classifications, reference [16] used 16 pretrained CNNs for identifying Covid positive and negative patients whereas reference [22] used seven Deep Neural Network (DNN) models. In both cases DenseNet performed the best. Reference [23] developed a deep neural network, CV19-Net that differentiated 2060 Covid pneumonia from 3148 nonCovid pneumonia on CXR. They claimed that the AI algorithm (AUC-0.94) performed better than experienced thoracic radiologists (AUC-0.85). To aid non-radiologist physicians and to decrease inter reader variability among radiologists in CXR interpretations of Covid related pneumonia, reference [24] experimented on Computer Aided Design (CAD) systems. Other binary Covid and nonCovid classification works have been given in references [6, 25, 26].
Among works of four class classification, [27, 28] classified normal, bacterial, viral pneumonia, and Covid. Reference [29] implemented three binary classifications for the same. Reference [30] also did for the same classes, using the Monte-Carlo Drop weights Bayesian convolutional neural networks but used only 68 Covid cases. CXR images were classified into four classes of pneumonia, tuberculosis, Covid, normal, and further Covid CXR images were classified into mild, medium, and severe based on severity by [31].
Reference [32] implemented incremental learning where the CXR images are initially checked for positive case of pneumonia and in the next stage the platform makes decision if it is Covid or another type of pneumonia. A pneumonia centric work in [17] stated that it had lesser chances of mis predicting pneumonia compared to normal cases and Covid-19. Reference [19] developed a tool for viral pneumonia detection using Confidence-Aware Anomaly Detection with a large dataset containing 18,774 healthy controls, 18,619 nonviral pneumonia, 5977 viral pneumonia cases. They made sure that all the cases of viral pneumonia were collected before the outbreak of Covid and attained an accuracy of 80.65%. A study on using ML and DL together was done by [33]. For obtaining a robust model with less data, in contrast to a transfer learning approach, SVM was used for classification of features obtained from a CNN model’s fully connected layer. Reference[34] proposed triple view CNN for extracting features from three lung views (left, right, overall) and attained an accuracy of 79.4% with Resnet50 by using 217 Covid cases only. When existing models were studied for Covid predictions in chest radiographs, using data before spread of Covid-19, these models were overfitted to their training and validation data and performed very poorly on new institutional dataset. An example of such a defect is seen in Covid-Net model [35]. When this model was tested on CXR images collected prior to Covid spread, it gave a high false-positive rate of up to 61% [13]. The sources of errors for overfitting were that the model focused on patient support devices attached to them or due to corelation with other findings. Another reason for the model bias could be that the appearance of Covid in CXR images are symmetric, peripheral-predominant nodular and ground glass opacities, but this appearance is found in other chest infections too [13].
Reference [36] summarized a list of domains (such as virus identification, statistical analysis, drug therapy, imaging and clinical prediction, population management and geographic tracing) where AI and big data technologies can be used for diagnosis, treatment, prevention, and management decision making. An app named; Predicts the Mortality of Covid-19 Patients (PMCP) was developed that predicts the mortality probability of Covid-19 patients when admitted to hospital [37]. Some prospective problems noticed during the literature survey are: unseen biases in dataset due to small dataset [3, 32, 37, 38]-[43], bias due to CXR acquisition settings, bias due to textual labelling in images (e.g., if all Covid images are from a particular dataset and normal images are from another dataset) etc. These biases might be used by DL models to discriminate between classes. If the test set is further smaller due to small dataset, then the statistical certainty on learning is not justifiable. The proposed work set forth an in-depth analysis using qualitative and quantitative assessment of CXR image classification. Table 1 shows previous works in literature, most of which, used lesser data per class or less covid samples or had visible class imbalance.
3 Methodology
The proposed Deep Learning models explained in this section follows the standard methods namely CXR data collection, image pre-processing, data splitting, augmentation, training, tuning, testing, and performance analysis. A block diagram explaining the proposed Deep learning models is shown in Fig. 1.
3.1 Dataset description
DL models that showed best performance on internal, narrow dataset failed to perform below standard on external dataset maybe due to variations in image processing, image acquisition protocols or overlooked pathology [13]. The clinical application and prediction of the infectious classes from CXR images is a task of critical nature due to the main reason that, this decision affects the further treatment procedure choices made by the clinicians. Hence the design of such deep learning models has to pass through multiple tests to validate that the classification task is based on relevant visual indicators and not improper information. To make sure of this factor, we have trained and tested the developed models on two datasets, each of which were gathered from different sources. Dataset 1 is Covid-19 radiography database from Kaggle and dataset 2 is Covid-19, Pneumonia and Normal Chest X-ray posteroanterior images from Mendeley data.
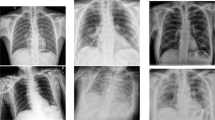
The CXR image samples used for the work have three classes labelled as Covid-19, normal, and pneumonia as shown in Fig. 2. Table 2 shows the patient case diversity in the dataset. To avoid the noticeable sparsity in the number of Covid-19 infected CXR images, like in most of the works in literature (Table 1), which caused significant bias, we have trained the model on 3 configurations of dataset. (a) a balanced dataset, DS1a (1345 images per class) which is a subset of DS1(b) an unbalanced dataset, DS1b (entire DS1) and (c) a balanced second dataset, DS2. The prediction model could be biased if it learns from an unbalanced dataset. The bias would be towards the class having more samples resulting in an inferior model for classification. So, we have used 2 sets of balanced (DS1a &DS2) and one unbalanced dataset (DS1b).
The DS1 in its 2nd update has 3616 Covid-19 positive cases, 10,192 Normal, 6012 Lung Opacity (Non-Covid lung infection), and 1345 Viral Pneumonia images. We have used only three categories i.e., Covid-19, normal, and pneumonia CXR images and dropped lung opacity for the experiments. The images of DS1 are in.png file format with a resolution of 299 by 299 pixels and a total of 15,153 samples.
The DS2 has Covid-19, Normal, and Pneumonia CXR posteroanterior images. It consists of 1525 images in each class and a total of 4575 samples. The images in DS2 are of varying resolutions and are in.png file format. DS1 consists of only viral pneumonia whereas DS2 has both viral and bacterial pneumonia cases.
3.2 Implementation of the deep learning model
The bias due to textual labelling, dataset variability, CXR acquisition settings etc., present in the images are mostly removed in the pre-processing stage, else the DL model perceives theses biases and directs the learning process. The datasets have different CXR image sizes, therefore based on the model requirements resizing is done. Every image in the curated dataset is passed through the image pre-processing pipeline that involves loading the image from the disk followed by image resizing, conversion of data and labelling into numpy array, the image normalization is done by scaling the pixel intensities to the range [0,1] and finally encoding categorical variables. During training, we have augmented the image batches in real time using ImageDataGenerator from the tensorflow.keras library, with rotation range of 15 for better learning and increased generalizing capability.
There are three successful techniques to use CNNs for medical image classification namely building a model from scratch, using the off the rack pretrained CNN features and doing unsupervised pretraining with supervised fine tuning [48]. We have adopted two strategies in the proposed work: one is developing two models by transfer learning and another by designing a CNN from scratch. In the first two works, we fine tune the last layer of a pretrained model. The basic steps involved in transfer learning are: using the pretrained model weights for the new model, including new layers for architecture modification of new model to make it adaptable to the problem at hand, new layers initialization, deciding which layers to be frozen and which layers to be put through the learning process, using a loss function and optimisation algorithm for weight updating and hence training [21].
Majority of the existing literature for Covid classification uses pretrained DNN. The limited Covid dataset is considered by these models which thus causes over fitting because the number of trainable parameters would then be larger than the amount of data at hand [49]. To rule out this scenario we have developed a CNN network from scratch too. The proposed three models are VGG16, IR, and CNN.
3.2.1 VGG16 Model
The transfer learning for this model was done by using weights of VGG16 available from pretraining on ImageNet dataset. The model head has the global average pooling layer (with pool size = (4,4)), a fully connected layer (with rectified linear unit-relu activation function) and a dropout layer (with a threshold = 0.5) that were added to the VGG16 base. For three class classification purpose, a Fully Connected (FC) layer having three units is added after the dropout layer (with softmax activation function). The actual model that will be trained is the head FC model. To make sure that the layers of the base model are not updated during the training process, we loop over all the layers in the base model and freeze them. Then we compile the model and train the network head. The hyper parameters that give optimized results for image resolution 224 by 224 are learning rate of 1e-3, 10 epochs and batch size of 32 with Adam optimizer and categorical cross entropy loss.
3.2.2 Inception Resnet V2 model
Using the InceptionResNetV2 keras model, we froze the weights of the top layer for transfer learning. The imagenet weights were used, with input image shape as 224 by 224 by 3 and classifier activation function set to sigmoid function. The global average pooling layer was followed by the final output layer, having softmax activation function. The primitive Inception-ResNetV2 network output has 1,000 classes. For our proposed work, only three classes are required: Covid-19, viral pneumonia, and normal. Therefore, the output channel number of the last layer (FC) is changed from 1000 to 3. The hyper parameters that give optimized results are learning rate of 1e-4, 10 epochs and batch size of 32 with adam optimizer and a custom defined loss function called focal loss.
Most of the previous works used dataset that has clear class imbalance as shown in Table 1. Practically the availability of Covid images is less, so data imbalance is an obvious scenario. When there is considerable data in one class, the majority class becomes well classified. Focal loss provides the well classified instances a lower weight, instead of providing all the training data with same weights [50]. So, focal loss gives the minority class instances more relative weight during training. This strategy to balance the weighing of the training instances helps when the dataset is unbalanced as in DS1b. Hence more training emphasis is placed on data that is difficult to classify.
3.2.3 CNN model from scratch
The architecture of a CNN consists of a number of sequenced layers. The layers and its tasks are as follows. The main building block is the convolution layer which revels the discriminative features of the input data by applying filters to the data. Following this layer is the nonlinearity/activation layer. Commonly used activation functions are sigmoid, Relu etc. Then comes the Pooling (Down-sampling) layer i.e., usually added to reduce the number of the computational nodes, between consecutive convolutional layers. Frequently used ones are maximum pooling, average pooling, and L2-norm pooling. Then the flatting layer collects the data in a single vector. And finally, the fully-connected layers ensure the connections between all activations and computational nodes. The CNN model summary is shown in Table 3.
The proposed CNN architecture has 3,609,955 trainable parameters in total. The fixed first layer of the sequential model is the input layer having an input shape of 299 by 299 by 3 pixels for DS1 and 500 by 500 by 3 for DS2. The remaining layers are functions of convolution, Relu, and max pooling layers. The last layer is the fully connected layer with the softmax function. The hyper parameters that gave the optimized results were learning rate of 1e-4 for DS1 and 1e-3 for DS2, number of epochs as 10 and batch size of 32 for DS1 and 128 for DS2 with adam optimizer and a custom defined loss function called focal loss. The focal loss is used as the loss function to address class imbalance. When using focal loss, the samples near the decision boundary are focused while the well classified sample’s weight was reduced.
4 Experimental setup
All the models were built and experimented using keras deep learning library with tensorflow backend. The batch size, number of epochs and learning rate are initialised. The images are then loaded from the dataset directory and the class labels are extracted from the file name. The images are pre-processed and are split into 80% as training set, 10% as validation set (both for training the model) and 10% as test sets (for evaluating the model). The model is compiled and training process is initiated. The deep learning algorithm teaches the patterns in input using the train set, by reducing the error between target outputs and predicted output. Adam optimizer is used to optimize the deep learning model weights and decrease the categorical cross entropy loss function. The trained model’s performance is then evaluated using the test set. The model is tested on a set of data that were not used in the training and validation process.
One class might over weigh the other classes, when a class is underrepresented, misleading to high classification accuracy. In order to avoid strong class imbalance, we used a subset of dataset one i.e., balanced (DS1b) and experimented. To combat overfitting multiple strategies have been employed such as dropout regularization, L2 weight regularization and data augmentation. We experimented all the three models with various optimizers (adam, adagrad, stochastic gradient descent, RMSprop, and adadelta), activation functions (sigmoid, Relu, and softmax), loss functions (sparse categorical cross entropy and categorical cross entropy). The experimental results of each trial with varying hyper parameters are listed in Table 4. The incremental learning approach of using small subsets of data (batches), eliminates the load of fitting the whole dataset into the memory. All experiments were performed on a PC with Windows 10 Pro (Intel® Xeon® CPU E5-2620 v4 @ 2.10 GHz, 64 GB RAM) using Spyder (4.2.5) IDE and programming language as Python (version 3.8.8). The deep learning frameworks used were keras with tensorflow backend.
5 Results
To scrutinize the developed models in a quantitative manner, we enumerated the train-validation-test accuracies, sensitivity, specificity, and F1 score using two datasets. The training set accuracy- loss curves and that of the validation set, after each epoch was monitored for various experiments. All the proposed models show a modest learning progression during the training time resulting in a decrease of both the train and validation loss, as shown in Fig. 3. In addition to overall model performance, individual class performance was also evaluated using the same metrics. The precision and recall metrics serves for performance predictions when the dataset is imbalanced [51]. A detailed class wise precision, recall, and F1 score is mentioned in the Additional file 1. All the tested models showed superior discrimination capabilities for Covid classes. To understand the model performance at a deeper level we studied the confusion matrix of each model. The confusion matrix of each and every simulation is evaluated and shown in Fig. 3. The mathematical values of the model’s performance metrics are shown in Fig. 4, along with the dataset and the models used. The networks are tested with hyperparameter values that gave the best performance on train and validation dataset. The proposed network outperformed majority of the literature works.
6 Model verification using GRAD-CAM
Any predictions made by a deep learning model (usually referred to as the black box classification) will be scrutinized by the healthcare providers in real time, despite developing an experimentally and highly accurate model. Reference [58] evaluated the performance of trained DL networks in hospital systems. They concluded that a high performing AI system got its performance from undesired shortcuts and hence AI systems should be verified whether they rely on the proper signals. To avoid this situation of shortcut learning, improved quality data must be used for designing a robust model. In the work presented in reference [58] Covid positive and negative was learned by the model, with positioning of shoulder, which was not a factor for decision making. This was a shortcut learning adopted by the DL model. The poor generalization performance is due to the choice of model architecture, the use of dataset from a single source etc. Large generalization gap was mostly due to shortcut learning [58].
Considering some of the state-of-the-art explainability models, the experimental results in [59] shows, the impact on decision making by LIME to be 38%, SHAP 44%, Expected Gradients 51% and GSInquire 76%. Even though these methods are not perfect they help in improving the trust on DL models which are often perceived as ‘black box’. Transparency and interpretability of model’s decision, is a necessity in medical imaging DL applications [59]. The Grad-CAM technique can be used to provide the explainable visualization of the DL models [60]. The proposed work utilised an open dataset, making it clear that open datasets are prone to heavy bias. The image quality could be varying because of being collected from different sources or noisy or have visible devices attached to ICU patients. Figure 5 shows the heat map responses generated by the Grad-CAM method of a Covid and pneumonia image each. The original image is in row 1 and row 2 shows, the heat map visualization (an overlay of heatmap onto the original image) highlighting the lung regions contributing to prediction. The heat map clearly shows that the model has focused on both sides of the lungs. It was noticed by the clinician that the probes visibility in some Xray’s were learned by the DL model as features causing bad learning behaviour. This was also revealed by the thermograms. These images were removed or cleaned before using for learning after suggestions by clinicians.
7 Discussion
The proposed study is based on the design of a classifier that could make one of the following decisions from a CXR image: (a) COVID-19 viral infection, (b) pneumonia infection, (c) normal (no infection). These predictions aid the medics to direct the patients towards RTPCR testing for Covid confirmations or decide on which patients to be prioritized for immediate treatment.
Using a balanced dataset with 1345 images per class i.e., a subset of dataset 1, VGG 16, IR, and CNN gave 96, 97, 97 percent accuracies respectively. With the whole dataset 1 i.e., a unbalanced dataset with a total of 15,153 CXR images, VGG 16, IR, and CNN gave 89,89, 96 percent accuracies respectively. With the dataset 2, also balanced with 1525 images per class, VGG 16, IR, and CNN gave 89, 94, 93 percent accuracies respectively. The best proposed model in terms of accuracy is CNN for all the dataset, as shown in Table 5. A sensitivity of 98.21, specificity of 96.62 and a F1 score of 98 was obtained for the best proposed model (CNN) whereas the highest attained sensitivity was 98.5, specificity of 97.28 and F1 score of 99 for IR model. IR model also performed almost equivalent to CNN but the training time for IR was high as 9 h as in Table 4. A reason for this is that the IR model has complex internal structured modules.
The model achieved a good balance between performance and computational efficiency. The notion that pretrained models dictate the image rescaling to 256 by 256 or 224 by 224 is overruled in the proposed CNN model, because we used 500 by 500 resolutions. On conducting a survey on all the deep learning models developed for the purpose of classification into Covid, normal, and pneumonia cases using CXR images, we compared the proposed model with a set of existing studies in terms of computation efficiency and performance in Table 6. It is noted that studies like [3, 12, 21, 26, 33, 38,39,40,41,42,43, 49, 54,55,56, 61,62,63] used remarkably smaller dataset for Covid images as shown in Table 1. This caused the model to lean more towards the class with higher number of images causing a major bias due to unbalanced images per class. Some high accuracy literature works have their flaws in using small dataset, low sensitivity, and high false negative. Some have low accuracy with less false predictions. Literature has classifications from 10 to 2 classes which include Covid-19 images but retained to using pretrained models only [64, 65].
According to reference [66] a model is prone to higher bias risk when it is trained on Covid positive images less than 500 and lower risk when 2000 Covid positive images are used. Hence, we have made sure to use 3616 Covid positive images in DS1b. Reference [66]concluded that none of the models he studied were of potential clinical application due to defects in methodology or inherent biases. Some reasons stated were poor integration of imaging data, bias in small dataset, dataset variability, difficulty of the prediction task and lack of teamwork by data analyst and clinicians so as to make the model implementable and clinically relevant. Suggestions given by this work, to solve these problems and develop higher quality models were considered in the proposed work. According to requirements to be satisfied by a DL model, we performed internal and external validation to give insight into algorithm’s generalizability, the robustness of our model was analysed by changing the seed value, data partition demographics are stated, mentioned limitations and addressed bias problems. It is important to note that the proposed networks are trained and tested on multiple datasets and hence it generalizes well for new cases.
8 Conclusion
The proposed work has some limitations. The model cannot determine the stage of the Covid-19 pneumonia. When comparing models over cross dataset and intra dataset, the performance is affected due to the data acquisition diversity. Over fitting due to narrow dataset has been addressed in the work but overfitting can be due to other broader problems. This could be solved by using a diverse selection of data from various disease types, multiple sites/sources and being precise about the population of patients being evaluated (i.e., ICU vs ED patients). As we have developed the models using public datasets and the characteristics of these data may be different from clinical data, we have to further work with clinical data. Future upgradations include improvement of sensitivity and PPV as new data accumulates over time. Further designs that can be integrated into the decision support systems may include predicting patient risk status, survival analysis, disease stage, prediction of recovery duration, as it would help in scenarios of fewer patients accommodating the hospital facilities.
Data availability
The datasets experimented and analysed during the current study are available in the below web links. Dataset1:https://www.kaggle.com/tawsifurrahman/Covid19-radiography-database. Dataset2: https://data.mendeley.com/datasets/jctsfj2sfn/1.
Code availability
The codes generated during the current study are available from the corresponding author upon reasonable request.
References
Rabi FA, Al Zoubi MS, Al-Nasser AD, et al. Sars-cov-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:1–14. https://doi.org/10.3390/pathogens9030231.
Chang L, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 2020;34:75–80. https://doi.org/10.1016/j.tmrv.2020.02.003.
Loey M, Smaarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep. Symmetry (Basel). 2020;12:651. https://doi.org/10.3390/sym12040651.
Singhal T. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatricshe Indian J Pediatrics. 2020;87:281–6.
Cleverley J, Piper J, Jones MM. The role of chest radiography in confirming covid-19 pneumonia. BMJ. 2020. https://doi.org/10.1136/bmj.m2426.
Minaee S, Kafieh R, Sonka M, et al. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. 2021;65:1–9. https://doi.org/10.1016/j.media.2020.101794.
Yang Y, Yang M, Yuan J, et al. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation. 2020;1:100061. https://doi.org/10.1016/j.xinn.2020.100061.
Ghafouri-Fard S, Noroozi R, Vafaee R, Branicki W. Effects of host genetic variations on response to, susceptibility and severity of respiratory infections. Biomed Pharmacother J. 2020;128:110296. https://doi.org/10.1016/j.biopha.2020.110296.
Ying S, Zheng S, Li L, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans Comput Biol Bioinform. 2020. https://doi.org/10.1101/2020.02.23.20026930.
Mohammad-Rahimi H, Nadimi M, Ghalyanchi-Langeroudi A, et al. Application of machine learning in diagnosis of COVID-19 through X-ray and CT images: a scoping review. Front Cardiovasc Med. 2021. https://doi.org/10.3389/fcvm.2021.638011.
Shorfuzzaman M, Masud M, Alhumyani H, et al. Artificial neural network-based deep learning model for COVID-19 patient detection using X-ray chest images. J Healthc Eng. 2021;2021:16. https://doi.org/10.1155/2021/5513679.
Alruwaili M, Shehab A, Abd El-Ghany S. COVID-19 diagnosis using an enhanced inception-ResNetV2 deep learning model in CXR images. J Healthc Eng. 2021;2021:16. https://doi.org/10.1155/2021/6658058.
Banerjee I, Sinha P, Purkayastha S, Mashhaditafreshi N, Tariq A, Jeong J, Trivedi H, Gichoya JW. Was there COVID-19 back in 2012? challenge for AI in diagnosis with similar indications. arXiv. 2020;2006:13262.
Tartaglione E, Barbano CA, Berzovini C, et al. Unveiling COVID-19 from chest x-ray with deep learning: a hurdles race with small data. Int J Environ Res Public Health. 2020;17:1–17. https://doi.org/10.3390/ijerph17186933.
Güneyli S, Atçeken Z, Doğan H, et al. Radiological approach to COVID-19 pneumonia with an emphasis on chest CT. Diagn Interv Radiol. 2020;26:323–32. https://doi.org/10.5152/dir.2020.20260.
Pham TD. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci Rep. 2020;10:1–8. https://doi.org/10.1038/s41598-020-74164-z.
Hasan MDK, Ahmed S, Abdullah ZME, et al. Deep learning approaches for detecting pneumonia in COVID-19 patients by analyzing chest X-ray images. Math Probl Eng. 2021;2021:8. https://doi.org/10.1155/2021/9929274.
Bai HX, Hsieh B, Xiong Z, et al. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296:E46–54. https://doi.org/10.1148/radiol.2020200823.
Zhang J, Xie Y, Pang G, et al. Viral pneumonia screening on chest X-rays using confidence-aware anomaly detection. IEEE Trans Med Imaging. 2021;40:879–90. https://doi.org/10.1109/TMI.2020.3040950.
Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–8. https://doi.org/10.1016/j.jare.2020.03.005.
Luz E, Silva P, Silva R, et al. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res Biomed Eng. 2021. https://doi.org/10.1007/s42600-021-00151-6.
Hemdan EE-D, Shouman MA, Karar ME COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. 2020.
Zhang R, Tie X, Qi Z, et al. Diagnosis of Coronavirus disease 2019 pneumonia by using chest radiography: value of artificial intelligence. Radiology. 2021;298:E88–97. https://doi.org/10.1148/RADIOL.2020202944.
Hwang EJ, Kim KB, Kim JY, et al. COVID-19 pneumonia on chest X-rays: Performance of a deep learning-based computer-aided detection system. PLoS ONE. 2021;16:1–16. https://doi.org/10.1371/journal.pone.0252440.
Sahlol AT, Yousri D, Ewees AA, et al. COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci Rep. 2020;10:1–15. https://doi.org/10.1038/s41598-020-71294-2.
Ozturk T, Talo M, Azra E, et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. https://doi.org/10.1016/j.compbiomed.2020.103792.
Ibrahim AU, Ozsoz M, Serte S, et al. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cognit Comput. 2021. https://doi.org/10.1007/s12559-020-09787-5.
Afshar P, Heidarian S, Naderkhani F, et al. COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit Lett. 2020;138:638–43. https://doi.org/10.1016/j.patrec.2020.09.010.
Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24:1207–20. https://doi.org/10.1007/s10044-021-00984-y.
Ghoshal B, Tucker A. Estimating Uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. ArXiv. 2020;2003:10769.
Shelke A, Inamdar M, Shah V, et al. Chest X-ray classification using deep learning for automated COVID-19 screening. SN Comput Sci. 2021. https://doi.org/10.1007/s42979-021-00695-5.
Yeh C-F, Cheng H-T, Wei A, et al. A cascaded learning strategy for robust COVID-19 pneumonia chest X-ray screening. 1–14. 2020.
Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Mathematical Eng Manag Sci. 2020;5:643–51. https://doi.org/10.33889/IJMEMS.2020.5.4.052.
Zhang J, Zhou L, Wang L. Triple-view Convolutional Neural Networks for COVID-19 Diagnosis with Chest X-ray. 1–10. 2020.
Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10:1–12. https://doi.org/10.1038/s41598-020-76550-z.
Dong J, Wu H, Zhou D, et al. Application of big data and artificial intelligence in COVID-19 prevention, diagnosis, treatment and management decisions in China. J Med Syst. 2021;45:1–11. https://doi.org/10.1007/s10916-021-01757-0.
Lin J-K, Chien T-W, Wang L-Y, Chou W. An artificial neural network model to predict the mortality of COVID-19 patients using routine blood samples at the time of hospital admission. Medicine. 2021;100:e26532. https://doi.org/10.1097/md.0000000000026532.
Zargari Khuzani A, Heidari M, Shariati SA. COVID-Classifier: an automated machine learning model to assist in the diagnosis of COVID-19 infection in chest X-ray images. Sci Rep. 2021;11:1–6. https://doi.org/10.1038/s41598-021-88807-2.
Li X, Li C, Zhu D. COVID-MobileXpert: On-Device COVID-19 Patient Triage and Follow-up using Chest X-rays. Proceedings—2020 IEEE International Conference on Bioinformatics and Biomedicine, BIBM. 2020;1063–1067. https://doi.org/10.1109/BIBM49941.2020.9313217
Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020. https://doi.org/10.1016/j.compbiomed.2020.103805.
Yildirim M, Cinar A. A deep learning based hybrid approach for COVID-19 disease detections. Traitement du Signal. 2020;37:461–8. https://doi.org/10.18280/ts.370313.
Novitasari DCR, Hendradi R, Caraka RE, et al. Detection of COVID-19 chest x-ray using support vector machine and convolutional neural network. Commun Math Biol Neurosci. 2020. https://doi.org/10.28919/cmbn/4765.
Civit-Masot J, Luna-Perejón F, Morales MD, Civit A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci. 2020;10:4640. https://doi.org/10.3390/app10134640.
Zhang Y, Niu S, Qiu Z, et al. COVID-DA: Deep domain adaptation from typical pneumonia to COVID-19. 1–8. 2020.
Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20:100412. https://doi.org/10.1016/j.imu.2020.100412.
Chowdhury MEH, Rahman T, Khandakar A, et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–76. https://doi.org/10.1109/ACCESS.2020.3010287.
Albahli S, Albattah W. Detection of coronavirus disease from X-ray images using deep learning and transfer learning algorithms. J Xray Sci Technol. 2020;28:841–50. https://doi.org/10.3233/XST-200720.
Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–98. https://doi.org/10.1109/TMI.2016.2528162.
Khobahi S, Agarwal C, Schonfeld D, Soltanalian M. CoroNet: a deep network architecture for enhanced identification of COVID-19 from chest x-ray images. 2021. https://doi.org/10.1117/12.2580738
Lin TY, Goyal P, Girshick R, et al. Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2020;42:318–27. https://doi.org/10.1109/TPAMI.2018.2858826.
Arora V, Ng EYK, Leekha RS, et al. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput Biol Med. 2021;135:104575. https://doi.org/10.1016/j.compbiomed.2021.104575.
Nishio M, Noguchi S, Matsuo H, Murakami T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: combination of data augmentation methods. Sci Rep. 2020;10:1–6. https://doi.org/10.1038/s41598-020-74539-2.
Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–40. https://doi.org/10.1007/s13246-020-00865-4.
Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform Med Unlocked. 2020;19:100360. https://doi.org/10.1016/j.imu.2020.100360.
Pandit MK, Banday SA. SARS n-CoV2-19 detection from chest x-ray images using deep neural networks. Int J Pervasive Comput Commun. 2020;16:419–27. https://doi.org/10.1108/IJPCC-06-2020-0060.
Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020;140:110122. https://doi.org/10.1016/j.chaos.2020.110122.
Rahaman MM, Li C, Yao Y, et al. Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J Xray Sci Technol. 2020;28:821–39. https://doi.org/10.3233/XST-200715.
DeGrave AJ, Janizek JD, Lee SI. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat Mach Intell. 2021;3:610–9. https://doi.org/10.1038/s42256-021-00338-7.
Lin ZQ, Shafiee MJ, Bochkarev S, Jules MS, Wang XY, Wong A. Do Explanations Reflect Decisions? A Machine-centric Strategy to Quantify the Performance of Explainability Algorithms. arXiv. 2019;1910:07387.
Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2020;128:336–59. https://doi.org/10.1007/s11263-019-01228-7.
Shibly KH, Dey SK, Islam MTU, Rahman MM. COVID faster R-CNN: a novel framework to diagnose novel coronavirus disease (COVID-19) in X-ray images. Inform Med Unlocked. 2020;20:100405. https://doi.org/10.1016/j.imu.2020.100405.
Narayan Das N, Kumar N, Kaur M, et al. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM. 2020. https://doi.org/10.1016/j.irbm.2020.07.001.
Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl Soft Comput. 2020;97:106580. https://doi.org/10.1016/j.asoc.2020.106580.
Javed Mehedi Shamrat FM, Azam S, Karim A, et al. LungNet22: a fine-tuned model for multiclass classification and prediction of lung disease using X-ray images. J Pers Med. 2022. https://doi.org/10.3390/jpm12050680.
Akter S, Shamrat FMJM, Chakraborty S, et al. Covid-19 detection using deep learning algorithm on chest X-ray images. Biology (Basel). 2021. https://doi.org/10.3390/biology10111174.
Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3:199–217. https://doi.org/10.1038/s42256-021-00307-0.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. G.M. has written the manuscript, developed the methodology, carried out the experiments, data curation and the simulations. M.S. has formulated the concept and supervised this work. S.B. and S.S. have provided the medical inputs and revisions. All the authors made critical revisions and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohan, G., Subashini, M.M., Balan, S. et al. A multiclass deep learning algorithm for healthy lung, Covid-19 and pneumonia disease detection from chest X-ray images. Discov Artif Intell 4, 20 (2024). https://doi.org/10.1007/s44163-024-00110-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44163-024-00110-x