Abstract
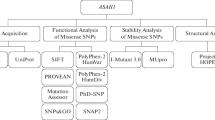
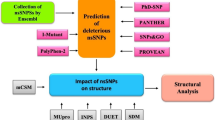
Niemann-Pick disease type C1 (NPC1), caused by mutations of NPC1 gene, is an inherited lysosomal lipid storage disorder. Loss of functional NPC1 causes the accumulation of free cholesterol (FC) in endocytic organelles that comprised the characteristics of late endosomes and/or lysosomes. In this study we analyzed the pathogenic effect of 103 nsSNPs reported in NPC1 using computational methods. R1186C, S940L, R958Q and I1061T mutations were predicted as most deleterious and disease associated with NPC1 using SIFT, Polyphen 2.0, PANTHER, PhD-SNP, Pmut and MUTPred tools which were also endorsed with previous in vivo experimental studies. To understand the atomic arrangement in 3D space, the native and disease associated mutant (R1186C, S940L, R958Q and I1061T) structures were modeled. Quantitative structural and flexibility analysis was conceded to observe the structural consequence of prioritized disease associated mutations (R1186C, S940L, R958Q and I1061T). Accessible surface area (ASA), free folding energy (FFE) and hydrogen bond (NH bond) showed more flexibility in 3D space in mutant structures. Based on the quantitative assessment and flexibility analysis of NPC1 variants, I1061T showed the most deleterious effect. Our analysis provides a clear clue to wet laboratory scientists to understand the structural and functional effect of NPC1 gene upon mutation.
Similar content being viewed by others
References
Adzhubei I A, Schmidt S, Peshkin L, Ramensky V E, Gerasimova A, Bork P, Kondrashov A S, Sunyaev S R (2010). A method and server for predicting damaging missense mutations. Nat Methods, 7(4): 248–249
Amberger J, Bocchini C A, Scott A F, Hamosh A (2009). McKusick’s online Mendelian inheritance in man (OMIM®). Nucleic Acids Res, 37(Database issue suppl 1): D793–D796
Balu K, Purohit R (2013). Mutational analysis of TYR gene and its structural consequences in OCA1A. Gene, 513(1): 184–195
Balu K, Rajendran V, Sethumadhavan R, Purohit R (2013). Investigation of binding phenomenon of NSP3 and p130Cas mutants and their effect on cell signalling. Cell Biochem Biophys, 67(2): 623–633
Bauer P, Knoblich R, Bauer C, Finckh U, Hufen A, Kropp J, Braun S, Kustermann-Kuhn B, Schmidt D, Harzer K, Rolfs A (2002). NPC1: Complete genomic sequence, mutation analysis, and characterization of haplotypes. Hum Mutat, 19(1): 30–38
Beltroy E P, Richardson J A, Horton J D, Turley S D, Dietschy J M (2005). Cholesterol accumulation and liver cell death in mice with Niemann-Pick type C disease. Hepatology, 42(4): 886–893
Capriotti E, Calabrese R, Casadio R (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics, 22(22): 2729–2734
Carstea E D, Morris J A, Coleman K G, Loftus S K, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman D B, Nagle J, Polymeropoulos M H, Sturley S L, Ioannou Y A, Higgins M E, Comly M, Cooney A, Brown A, Kaneski C R, Blanchette-Mackie E J, Dwyer N K, Neufeld E B, Chang T Y, Liscum L, Strauss J F 3rd, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O’Neill R R, van Diggelen O P, Elleder M, Patterson M C, Brady R O, Vanier M T, Pentchev P G, Tagle D A (1997). Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science, 277(5323): 228–231
Carvalho M A, Marsillac S M, Karchin R, Manoukian S, Grist S, Swaby R F, Urmenyi T P, Rondinelli E, Silva R, Gayol L, Baumbach L, Sutphen R, Pickard-Brzosowicz J L, Nathanson K L, Sali A, Goldgar D, Couch F J, Radice P, Monteiro A N A (2007). Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res, 67(4): 1494–1501
Carvalho M, Pino M A, Karchin R, Beddor J, Godinho-Netto M, Mesquita R D, Rodarte R S, Vaz D C, Monteiro V A, Manoukian S, Colombo M, Ripamonti C B, Graeme Suthers R R, Borg A, Radice P, Grist S A, Monteiro A N A, Billack B (2009). Analysis of a set of missense, frameshift, and in-frame deletion variants of BRCA1. Mutat Res, 660(1): 1–11
Chasman D, Adams RM (2001). Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol, 307(2): 683–706
Fernandez-Valero EM, Ballart A, Iturriaga C, Lluch M, Macias J, Vanier M T, Pineda M, Coll M J (2005). Identification of 25 new mutations in 40 unrelated Spanish Niemann-Pick type C patients: genotypephenotype correlations. Clin Genet, 68(3): 245–254
Ferrer-Costa C, Gelpí J L, Zamakola L, Parraga I, de la Cruz X, Orozco M(2005). PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics, 21(14): 3176–3178
Garver W S, Francis G A, Jelinek D, Shepherd G, Flynn J, Castro G, Walsh Vockley C, Coppock D L, Pettit K M, Heidenreich R A, Meaney F J (2007). The National Niemann-Pick C1 disease database: report of clinical features and health problems. Am J Med Genet A, 143A(11): 1204–1211
Garver WS, Heidenreich R A (2002). The Niemann-Pick C proteins and trafficking of cholesterol through the late endosomal/lysosomal system. Curr Mol Med, 2(5): 485–505
Garver W S, Jelinek D, Meaney F J, Flynn J, Pettit K M, Shepherd G, Heidenreich R A, Vockley C M W, Castro G, Francis G A (2010). The National Niemann-Pick Type C1 Disease Database: correlation of lipid profiles, mutations, and biochemical phenotypes. J Lipid Res, 51(2): 406–415
Goldgar D E, Easton D F, Deffenbaugh AM, Monteiro A N, Tavtigian S V, Couch F J, the Breast Cancer Information Core (BIC) Steering Committee (2004). Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet, 75(4): 535–544
Goldstein J L, Brown M S (1992). Lipoprotein receptors and the control of plasma LDL cholesterol levels. Eur Heart J, 13(Suppl B): 34–36
Greer W L, Dobson M J, Girouard G S, Byers D M, Riddell D C, Neumann P E (1999). Mutations in NPC1 highlight a conserved NPC1-specific cysteine-rich domain, Am J Hum Genet, 65(5): 1252–1260
Harpaz Y, Gerstein M, Chothia C (1994). Volume changes on protein folding. Structure, 2(7): 641–649
Hess B, Kutzner C, Van Der Spoel D, Lindahl E (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput, 4(3): 435–447
Hollup S M, Salensminde G, Reuter N (2005). WEBnm@: a web application for normal mode analyses of proteins. BMC Bioinformatics, 6(1): 52
Ioannou Y A (2000). The structure and function of the Niemann-Pick C1 protein. Mol Genet Metab, 71(1–2): 175–181
Kamaraj B, Purohit R (2014). Computational screening of disease-associated mutations in OCA2 gene. Cell Biochem Biophys, 68(1): 97–109
Kamaraj B, Purohit R (2013). In silico screening and molecular dynamics simulation of disease-associated nssnp in tyrp1 gene and its structural consequences in OCA3. BioMed Res Int, 2013: Article ID 697051
Kaplan W, Littlejohn T G (2001). Swiss-PDB viewer (deep view). Brief Bioinform, 2(2): 195–197
Karchin R (2009). Next generation tools for the annotation of human SNPs. Brief Bioinform, 10(1): 35–52
Kobayashi T, Beuchat M H, Lindsay M, Frias S, Palmiter R D, Sakuraba H, Parton R G, Gruenberg J (1999). Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol, 1(2): 113–118
Kumar A, Purohit R (2012a). Computational centrosomics: an approach to understand the dynamic behaviour of centrosome. Gene, 511(1): 125–126
Kumar A, Purohit R (2012c). Computational investigation of pathogenic nsSNPs in CEP63 protein. Gene, 503(1): 75–82
Kumar A, Purohit R (2013). Cancer associated E17K mutation causes rapid conformational drift in AKT1 pleckstrin homology (PH) domain. PLoS One, 8(5): e64364
Kumar A, Purohit R (2014). Use of Long Term Molecular Dynamics Simulation in Predicting Cancer Associated SNPs. PLoS Comput Biol, 10(4): e1003318
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2013). Molecular dynamic simulation reveals damaging impact of RAC1 F28L mutation in the switch I region. PLoS One, 8(10): e77453
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2013a). Computational investigation of cancer-associated molecular mechanism in Aurora A (S155R) mutation. Cell Biochem Biophys, 66(3): 787–796
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2013b). Evidence of colorectal cancer-associated mutation in MCAK: a computational report. Cell Biochem Biophys, 67(3): 837–851
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2014). Relationship between a point mutation S97C in CK1δ protein and its affect on ATP-binding affinity. J Biomol Struct Dyn, 32(3): 394–405
Kumar A, Rajendran V, Sethumadhavan R, Shukla P, Tiwari S, Purohit R (2014). Computational SNP analysis: current approaches and future prospects. Cell Biochem Biophys, 68(2): 233–239
Kumar P, Henikoff S, Ng P C (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc, 4(7): 1073–1081
Kumar A, Purohit R (2012b). Computational screening and molecular dynamics simulation of disease associated nsSNPs in CENP-E. Mutat Res, 738-739: 28–37
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2012). In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ). FEBS Open Bio, 2: 285–293
Laskowski R A, Rullmannn J A, MacArthur M W, Kaptein R, Thornton J M (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR, 8(4): 477–486
Li B, Krishnan V G, Mort M E, Xin F, Kamati K K, Cooper D N, Mooney S D, Radivojac P (2009). Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics, 25(21): 2744–2750
Liscum L (2000). Niemann-Pick type C mutations cause lipid traffic jam. Traffic, 1(3): 218–225
Millat G, Baïlo N, Molinero S, Rodriguez C, Chikh K, Vanier M T (2005). Niemann-Pick C disease: use of denaturing high performance liquid chromatography for the detection of NPC1 and NPC2 genetic variations and impact on management of patients and families. Mol Genet Metab, 86(1–2): 220–232
Millat G, Marçais C, Rafi M A, Yamamoto T, Morris J A, Pentchev P G, Ohno K, Wenger D A, Vanier M T (1999). Niemann-Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet, 65(5): 1321–1329
Millat G, Marçais C, Tomasetto C, Chikh K, Fensom A H, Harzer K, Wenger D A, Ohno K, Vanier M T (2001). Niemann-Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am J Hum Genet, 68(6): 1373–1385
Mooney S (2005). Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform, 6(1): 44–56
Morris J A, Zhang D, Coleman K G, Nagle J, Pentchev P G, Carstea E D (1999). The genomic organization and polymorphism analysis of the human Niemann-Pick C1 gene. Biochem Biophys Res Commun, 261(2): 493–498
Ng P C, Henikoff S (2006). Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet, 7(1): 61–80
Pipalia N H, Huang A, Ralph H, Rujoi M, Maxfield F R (2006). Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J Lipid Res, 47(2): 284–301
Puri V, Watanabe R, Dominguez M, Sun X, Wheatley C L, Marks D L, Pagano R E (1999). Cholesterol modulates membrane traffic along the endocytic pathway in sphingolipid-storage diseases. Nat Cell Biol, 1(6): 386–388
Purohit R (2014). Role of ELA region in auto-activation of mutant KIT receptor: a molecular dynamics simulation insight. J Biomol Struct Dyn, 32(7): 1033–1046
Purohit R, Rajendran V, Sethumadhavan R (2011a). Studies on adaptability of binding residues and flap region of TMC-114 resistance HIV-1 protease mutants. J Biomol Struct Dyn, 29(1): 137–152
Purohit R, Rajendran V, Sethumadhavan R (2011b). Relationship between mutation of serine residue at 315th position in M. tuberculosis catalase-peroxidase enzyme and Isoniazid susceptibility: an in silico analysis. J Mol Model, 17(4): 869–877
Purohit R, Sethumadhavan R (2009). Structural basis for the resilience of Darunavir (TMC114) resistance major flap mutations of HIV-1 protease. Interdiscip Sci, 1(4): 320–328
Rajendran V, Purohit R, Sethumadhavan R (2012). In silico investigation of molecular mechanism of laminopathy caused by a point mutation (R482W) in lamin A/C protein. Amino Acids, 43(2): 603–615
Rajendran V, Sethumadhavan R (2014). Drug resistance mechanism of PncA in Mycobacterium tuberculosis. J Biomol Struct Dyn, 32(2): 209–221
Ribeiro I, Marcão A, Amaral O, Sá Miranda M C, Vanier M T, Millat G (2001). Niemann-Pick type C disease: NPC1 mutations associated with severe and mild cellular cholesterol trafficking alterations. Hum Genet, 109(1): 24–32
Runz H, Dolle D, Schlitter A M, Zschocke J (2008). NPC-db, a Niemann-Pick type C disease gene variation database. Hum Mutat, 29(3): 345–350
Scott C, Ioannou Y A (2004). The NPC1 protein: structure implies function. Biochimica et Biophysica Acta (BBA)-Mol Cell Biol L, 1685(1): 8–13
Sherry S T, Ward M H, Kholodov M, Baker J, Phan L, Smigielski E M, Sirotkin K (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res, 29(1): 308–311
Steward R E, MacArthur M W, Laskowski R A, Thornton J M (2003). Molecular basis of inherited diseases: a structural perspective. Trends Genet, 19(9): 505–513
Sun X, Marks D L, Park W D, Wheatley C L, Puri V, O’Brien J F, Kraft D L, Lundquist P A, Patterson M C, Pagano R E, Snow K (2001). Niemann-Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am J Hum Genet, 68(6): 1361–1372
Sun X, Marks D L, Park W D, Wheatley C L, Puri V, O’Brien J F, Kraft D L, Lundquist P A, Patterson M C, Pagano R E, Snow K (2001). Niemann-Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am J Hum Genet, 68(6): 1361–1372
Sunyaev S, Ramensky V, Bork P (2000). Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet, 16(5): 198–200
Tamura H, Takahashi T, Ban N, Torisu H, Ninomiya H, Takada G, Inagaki N (2006). Niemann-Pick type C disease: novel NPC1 mutations and characterization of the concomitant acid sphingomyelinase deficiency. Mol Genet Metab, 87(2): 113–121
Tarugi P, Ballarini G, Bembi B, Battisti C, Palmeri S, Panzani F, Di Leo E, Martini C, Federico A, Calandra S (2002). Niemann-Pick type C disease: mutations of NPC1 gene and evidence of abnormal expression of some mutant alleles in fibroblasts. J Lipid Res, 43(11): 1908–1919
Thomas P D, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res, 13(9): 2129–2141
Vanier M T, Suzuki K (1998). Recent advances in elucidating Niemann-Pick C disease. Brain Pathol, 8(1): 163–174
Wiederstein M, Sippl M J (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res, 35(Web Server issue suppl 2): W407–10
Willard L, Ranjan A, Zhang H, Monzavi H, Boyko R F, Sykes B D, Wishart D S (2003). VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res, 31(13): 3316–3319
Xiong H, Higaki K, Wei C J, Bao X H, Zhang Y H, Fu N, Qin J, Adachi K, Kumura Y, Ninomiya H, Nanba E, Wu X R (2012). Genotype/phenotype of 6 Chinese cases with Niemann-Pick disease type C. Gene, 498(2): 332–335
Yamamoto T, Ninomiya H, Matsumoto M, Ohta Y, Nanba E, Tsutsumi Y, Yamakawa K, Millat G, Vanieer M, Pentchev P G, Ohno K (2000). Genotype-phenotype relationship of Niemann-Pick disease type C: a possible correlation between clinical onset and levels of NPC1 protein in isolated skin fibroblasts. J Med Genet, 37(9): 707–712
Zhang Y (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9(1): 40
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kandakatla, N., Ramakrishnan, G., Chekkara, R. et al. Computational screening of disease associated mutations on NPC1 gene and its structural consequence in Niemann-Pick type-C1. Front. Biol. 9, 410–421 (2014). https://doi.org/10.1007/s11515-014-1314-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11515-014-1314-2