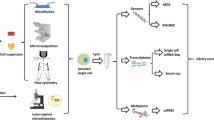
Abstract
The newly developed next-generation sequencing platforms, in combination with genome-scale amplification methods, provide a powerful tool to study genomics from a single cell. This mini-review summarizes the technologies of single cell genomics and their applications in several areas of biomedical research including stem cells, cancer biology and reproductive medicine. Particularly, it highlights recent advances in single cell exome sequencing, RNA-seq, and genome sequencing. The application of these powerful techniques will shed new light on the fundamental principles of gene transcription and genome organization at single-cell level and improve our understanding of cellular heterogeneity and diversity in multicellular organisms.
Similar content being viewed by others
References
Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe D B, Nusbaum C, Gnirke A (2011). Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol, 12(2): R18
Allen L Z, Ishoey T, Novotny M A, McLean J S, Lasken R S, Williamson S J (2011). Single virus genomics: a new tool for virus discovery. PLoS ONE, 6(3): e17722
Bhutani N, Burns D M, Blau H M (2011). DNA demethylation dynamics. Cell, 146(6): 866–872
Buganim Y, Faddah D A, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm S L, van Oudenaarden A, Jaenisch R (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell, 150(6): 1209–1222
Chaffer C L, Weinberg R A (2011). A perspective on cancer cell metastasis. Science, 331(6024): 1559–1564
Clark MJ, Homer N, O’Connor B D, Chen Z, Eskin A, Lee H, Merriman B, Nelson S F (2010). U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet, 6(1): e1000832
Corneveaux J J, Kruer M C, Hu-Lince D, Ramsey K E, Zismann V L, Stephan D A, Craig D W, Huentelman M J (2007). SNP-based chromosomal copy number ascertainment following multiple displacement whole-genome amplification. Biotechniques, 42(1): 77–83
Dalerba P, Kalisky T, Sahoo D, Rajendran P S, Rothenberg M E, Leyrat A A, Sim S, Okamoto J, Johnston D M, Qian D, Zabala M, Bueno J, Neff N F, Wang J, Shelton A A, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke M F, Quake S R (2011). Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol, 29(12): 1120–1127
Dean F B, Hosono S, Fang L, Wu X, Faruqi A F, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore S F, Egholm M, Lasken R S (2002). Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA, 99(8): 5261–5266
Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S (2009). Real-time DNA sequencing from single polymerase molecules. Science, 323(5910): 133–138
Esteller M (2007). Cancer epigenomics: DNA methylomes and histonemodification maps. Nat Rev Genet, 8(4): 286–298
Fan H C, Wang J, Potanina A, Quake S R (2011). Whole-genome molecular haplotyping of single cells. Nat Biotechnol, 29(1): 51–57
Farago C, Chester I C (1961). Cancer in the Territory of Papua and New Guinea: a preliminary communication. Med J Aust, 48(2): 1033–1035
Feng Z, Fang G, Korlach J, Clark T, Luong K, Zhang X, Wong W, Schadt E (2013). Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLOS Comput Biol, 9(3): e1002935
Frontera WR, Zayas A R, Rodriguez N (2012). Aging of human muscle: understanding sarcopenia at the single muscle cell level. Phys Med Rehabil Clin N Am, 23(1): 201–207, xiii
Galán A, Montaner D, Póo ME, Valbuena D, Ruiz V, Aguilar C, Dopazo J, Simón C (2010). Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS ONE, 5(10): e13615
Geschwind D H, Konopka G (2009). Neuroscience in the era of functional genomics and systems biology. Nature, 461(7266): 908–915
Guo Y, Yang Y, Zhou J, Czajkowsky D M, Liu B, Shao Z (2012). Microdissection of spatially identified single nuclei in a solid tumor for single cell whole genome sequencing. Biotechniques, 0(0): 1–3
Hamburger A W, Salmon S E (1977). Primary bioassay of human tumor stem cells. Science, 197(4302): 461–463
Hanson E K, Ballantyne J (2005). Whole genome amplification strategy for forensic genetic analysis using single or few cell equivalents of genomic DNA. Anal Biochem, 346(2): 246–257
Hashimshony T, Wagner F, Sher N, Yanai I (2012). CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Reports 2, 666–673
He C (2010). Grand challenge commentary: RNA epigenetics? Nat Chem Biol, 6(12): 863–865
Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann E M, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, Höfler G, Eisner F, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl J B, Speicher M R (2013). Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res, 73(10): 2965–2975
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, Wu H, Ye X, Ye C, Wu R, Jian M, Chen Y, Xie W, Zhang R, Chen L, Liu X, Yao X, Zheng H, Yu C, Li Q, Gong Z, Mao M, Yang X, Yang L, Li J, Wang W, Lu Z, Gu N, Laurie G, Bolund L, Kristiansen K, Wang J, Yang H, Li Y, Zhang X, Wang J (2012). Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell, 148(5): 873–885
Hughes S, Jones J L (2007). The use of multiple displacement amplified DNA as a control for methylation specific PCR, pyrosequencing, bisulfite sequencing and methylation-sensitive restriction enzyme PCR. BMC Mol Biol, 8(1): 91
Hutchison C A 3rd, Smith H O, Pfannkoch C, Venter J C (2005). Cellfree cloning using phi29 DNA polymerase. Proc Natl Acad Sci USA, 102(48): 17332–17336
Iourov I Y, Vorsanova S G, Yurov Y B (2012). Single cell genomics of the brain: focus on neuronal diversity and neuropsychiatric diseases. Curr Genomics, 13(6): 477–488
Islam S, Kjällquist U, Moliner A, Zajac P, Fan J B, Lönnerberg P, Linnarsson S (2011). Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res, 21(7): 1160–1167
Jones P A, Baylin S B (2007). The epigenomics of cancer. Cell, 128(4): 683–692
Kleinsmith L J, Pierce G B Jr (1964). Multipotentiality of Single Embryonal Carcinoma Cells. Cancer Res, 24: 1544–1551
Lecault V, Vaninsberghe M, Sekulovic S, Knapp D J, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, Falconnet D, White A K, Kent D G, Copley M R, Taghipour F, Eaves C J, Humphries R K, Piret J M, Hansen C L (2011). High-throughput analysis of single hematopoietic stem cell proliferation in micro-fluidic cell culture arrays. Nat Methods, 8(7): 581–586
Lee J Y, Dong S M, Kim S Y, Yoo N J, Lee S H, Park W S (1998). A simple, precise and economical microdissection technique for analysis of genomic DNA from archival tissue sections. Virchows Archiv: an international journal of pathology 433, 305–309
Li L, Clevers H (2010). Coexistence of quiescent and active adult stem cells in mammals. Science, 327(5965): 542–545
Ling J, Deng Y, Long X, Liu J, Du H, Cao B, Xu K (2012). Singlenucleotide polymorphism array coupled with multiple displacement amplification: accuracy and spatial resolution for analysis of chromosome copy numbers in few cells. Biotechnol Appl Biochem, 59(1): 35–44
Lu S, Zong C, Fan W, Yang M, Li J, Chapman A R, Zhu P, Hu X, Xu L, Yan L, Bai F, Qiao J, Tang F, Li R, Xie X S (2012). Probing meiotic recombination and aneuploidy of single sperm cells by wholegenome sequencing. Science, 338(6114): 1627–1630
Luthra R, Medeiros L J (2004). Isothermal multiple displacement amplification: a highly reliable approach for generating unlimited high molecular weight genomic DNA from clinical specimens. J Mol Diagn, 6(3): 236–242
Mardis E R (2008). The impact of next-generation sequencing technology on genetics. Trends Genet, 24(3): 133–141
McLean J S, Lombardo M J, Ziegler M G, Novotny M, Yee-Greenbaum J, Badger J H, Tesler G, Nurk S, Lesin V, Brami D, Hall A P, Edlund A, Allen L Z, Durkin S, Reed S, Torriani F, Nealson K H, Pevzner P A, Friedman R, Venter J C, Lasken R S (2013). Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res, 23(5): 867–877
McVean G A, Myers S R, Hunt S, Deloukas P, Bentley D R, Donnelly P (2004). The fine-scale structure of recombination rate variation in the human genome. Science, 304(5670): 581–584
McWilliam Leitch E C, McLauchlan J (2013). Determining the cellular diversity of hepatitis C virus quasispecies by single-cell viral sequencing. J Virol, 87(23): 12648–12655
Metzker M L (2010). Sequencing technologies-the next generation. Nat Rev Genet, 11(1): 31–46
Morozova O, Marra M A (2008). Applications of next-generation sequencing technologies in functional genomics. Genomics, 92(5): 255–264
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie W R, Hicks J, Wigler M (2011). Tumour evolution inferred by single-cell sequencing. Nature, 472(7341): 90–94
Niedringhaus T P, Milanova D, Kerby M B, Snyder M P, Barron A E (2011). Landscape of next-generation sequencing technologies. Anal Chem, 83(12): 4327–4341
Paez J G, Lin M, Beroukhim R, Lee J C, Zhao X, Richter D J, Gabriel S, Herman P, Sasaki H, Altshuler D, Li C, Meyerson M, Sellers W R (2004). Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res, 32(9): e71
Pan X, Durrett R E, Zhu H, Tanaka Y, Li Y, Zi X, Marjani S L, Euskirchen G, Ma C, Lamotte R H, Park I H, Snyder M P, Mason C E, Weissman S M (2013). Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc Natl Acad Sci USA, 110(2): 594–599
Pan X, Urban A E, Palejev D, Schulz V, Grubert F, Hu Y, Snyder M, Weissman SM (2008). A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc Natl Acad Sci USA, 105(40): 15499–15504
Park S Y, Gönen M, Kim H J, Michor F, Polyak K (2010). Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest, 120(2): 636–644
Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D (2012). The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Reports 2: 766–773
Ramsköld D, Luo S, Wang Y C, Li R, Deng Q, Faridani O R, Daniels G A, Khrebtukova I, Loring J F, Laurent L C, Schroth G P, Sandberg R (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol, 30(8): 777–782
Reya T, Morrison S J, Clarke M F, Weissman I L (2001). Stem cells, cancer, and cancer stem cells. Nature, 414(6859): 105–111
Rothberg J M, Hinz W, Rearick T M, Schultz J, Mileski W, Davey M, Leamon J H, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons J F, Marran D, Myers J W, Davidson J F, Branting A, Nobile J R, Puc B P, Light D, Clark T A, Huber M, Branciforte J T, Stoner I B, Cawley S E, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza J A, Namsaraev E, McKernan K J, Williams A, Roth G T, Bustillo J (2011). An integrated semiconductor device enabling non-optical genome sequencing. Nature, 475(7356): 348–352
Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno K D, Imai T, Ueda H R (2013). Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol, 14(4): R31
Schoenborn L, Yates P S, Grinton B E, Hugenholtz P, Janssen P H (2004). Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microbiol, 70(7): 4363–4366
Shalek A K, Satija R, Adiconis X, Gertner R S, Gaublomme J T, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta J J, Gennert D, Gnirke A, Goren A, Hacohen N, Levin J Z, Park H, Regev A (2013). Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature, 498(7453): 236–240
Shendure J, Ji H (2008). Next-generation DNA sequencing. Nat Biotechnol, 26(10): 1135–1145
Simone N L, Bonner R F, Gillespie J W, Emmert-Buck M R, Liotta L A (1998). Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet, 14(7): 272–276
Song C X, Clark T A, Lu X Y, Kislyuk A, Dai Q, Turner S W, He C, Korlach J (2012). Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods, 9(1): 75–77
Tahiliani M, Koh K P, Shen Y, Pastor W A, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu D R, Aravind L, Rao A (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324(5929): 930–935
Tan D W, Jensen K B, Trotter M W, Connelly J T, Broad S, Watt F M (2013). Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development, 140(7): 1433–1444
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch B B, Siddiqui A, Lao K, Surani MA (2009). mRNASeq whole-transcriptome analysis of a single cell. Nat Methods, 6(5): 377–382
Tang F, Lao K, Surani M A (2011). Development and applications of single-cell transcriptome analysis. Nat Methods, 8(4 Suppl): S6–S11
Torres L, Ribeiro F R, Pandis N, Andersen J A, Heim S, Teixeira M R (2007). Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat, 102(2): 143–155
Tzvetkov M V, Becker C, Kulle B, Nürnberg P, Brockmöller J, Wojnowski L (2005). Genome-wide single-nucleotide polymorphism arrays demonstrate high fidelity of multiple displacement-based whole-genome amplification. Electrophoresis, 26(3): 710–715
Unger M A, Chou H P, Thorsen T, Scherer A, Quake S R (2000). Monolithic microfabricated valves and pumps by multilayer soft lithography. Science, 288(5463): 113–116
Voet T, Kumar P, Van Loo P, Cooke S L, Marshall J, Lin M L, Zamani Esteki M, Van der Aa N, Mateiu L, McBride D J, Bignell G R, McLaren S, Teague J, Butler A, Raine K, Stebbings L A, Quail M A, D’Hooghe T, Moreau Y, Futreal P A, Stratton M R, Vermeesch J R, Campbell P J (2013). Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res, 41(12): 6119–6138
Wang J, Fan H C, Behr B, Quake S R (2012). Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell, 150(2): 402–412
Wu A R, Neff N F, Kalisky T, Dalerba P, Treutlein B, Rothenberg M E, Mburu F M, Mantalas G L, Sim S, Clarke M F, Quake S R (2013). Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, He W, Zeng L, Xing M, Wu R, Jiang H, Liu X, Cao D, Guo G, Hu X, Gui Y, Li Z, Xie W, Sun X, Shi M, Cai Z, Wang B, Zhong M, Li J, Lu Z, Gu N, Zhang X, Goodman L, Bolund L, Wang J, Yang H, Kristiansen K, Dean M, Li Y, Wang J (2012). Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell, 148(5): 886–895
Xue Z, Huang K, Cai C, Cai L, Jiang C Y, Feng Y, Liu Z, Zeng Q, Cheng L, Sun Y E, Liu J Y, Horvath S, Fan G (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature, 500(7464): 593–597
Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol, 20(9): 1131–1139
Zandi S, Ahsberg J, Tsapogas P, Stjernberg J, Qian H, Sigvardsson M (2012). Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc Natl Acad Sci USA, 109(39): 15871–15876
Zhao Y, Gong X, Chen L, Li L, Liang Y, Chen S, Zhang Y (2013). Sitespecific methylation of placental HSD11B2 gene promoter is related to intrauterine growth restriction. Eur J Hum Genet, doi: 10.1038/ejhg.2013.226
Zong C, Lu S, Chapman A R, Xie X S (2012). Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science, 338(6114): 1622–1626
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Wang, Q., Zhu, X., Feng, Y. et al. Single-cell genomics: An overview. Front. Biol. 8, 569–576 (2013). https://doi.org/10.1007/s11515-013-1285-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11515-013-1285-8