Abstract
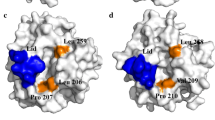
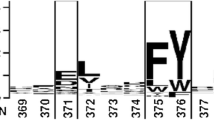
In order to improve the optimum temperature of lipases, the Penicillum expansum lipase (PEL) gene was mutated by site-directed mutagenesis using overlap extension PCR technique. The recombinant plasmid containing mutant E83V pPIC3.5K-lip-E83V was expressed in Pichia pastoris GS115. Comparison experiments of the mutant PEL-E83V-GS and the wild-type PEL-GS showed that the optimum temperature (45°C) of the mutant was 5°C higher than that of the wild type. The thermostability of the mutant was similar to that of the wild type. The enzymatic activity of the mutant was 188 U/ml at 37°C, which was 80% that of the wild type in the same conditions. Hydrophobic interaction may be enhanced in the surface region by the hydrophilic amino acid Glu substituted with the hydrophobic amino acid Val, and may be responsible for the improvement of the optimum temperature.
Similar content being viewed by others
References
Shi Q. Q., Study on alkaline lipase I. Screening and purification of the microorganism, Microbiology, 1981, 8: 109–110 (in Chinese)
Zheng Y., Huang J. Z., Shi Q. Q., We L. X. and Wu S. G., Study on alkaline-mesophile lipase III. Purification and some properties of alkaline lipase from Penicillum expansum PF868, Ind. Microbiol., 1996, 26(3): 15–18 (in Chinese)
Linn L., Xien B. F., Yang G. Z., Shi Q. Q., Lin Q. Y., Xie L. H., Wu S. G. and Wu X. F., Cloning and sequence analysis of cDNA encoding alkaline lipase from Penicillium expansumPF898, Chin. J. Chem. Mol. Biol., 2002, 18(1):32–37 (in Chinese)
Lin L., Xie B. F., Yang G. Z., Shi Q. Q., Lin Q. Y., Xie L. H., Wu S. G. and Wu X. F., Cloning and sequencing of genomic DNA encoding alkaline lipase from Penicillium expansum PF898, Chin. J. Chem. Mol. Biol., 2003, 19: 12–14 (in Chinese)
Yuan C. and Lin L., Overexpression of Penicillium expansum lipase gene in Pichia pastoris, Chin. J. Biotechnol., 2003, 19: 231–235 (in Chinese)
Beisson F., Tiss A., River C. and Verge R., Methods for lipase detection and assay, Eur. J. Lipid Technol., 2002, 2: 133–153
Kohno M., Enatsu M., Funatsu F., Yoshiizumi M. and Kugimiya W., Improvement of the optimum temperature of lipase activity for Rhizopus niveus by random mutagenesis and its structural interpretation, J. Biotechnol., 2001, 87: 203–210
Pack S. P. and Yoo Y. J., Protein thermostability: structure-based difference of residual properties between thermophilic and mesophilic proteins, J. Mol. Catal. B Enzym., 2003, 26: 257–264
Zhu G. P., Teng M. K. and Wang Y. Z., Contribution of proline to protein thermostability, Chin. J. Biotechnol., 2000, 20: 48–50 (in Chinese)
Watanabe K. and Suzuki Y., Protein thermostability by proline substitutions, J. Mol. Catal. B Enzym., 1998, 4: 167–180
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Microbiology, 2005, 32(1) (in Chinese)
Rights and permissions
About this article
Cite this article
Chen, G., Lin, L. Improvement of the Optimum Temperature of Penicillium expansum Lipase by Site-Directed Mutagenesis. Front. Biol. China 1, 1–4 (2006). https://doi.org/10.1007/s11515-005-0003-6
Issue Date:
DOI: https://doi.org/10.1007/s11515-005-0003-6