Abstract
Objective
Rational engineering of the crevice-like binding site of lipases for improvement of lipases’ catalytic properties.
Resuts
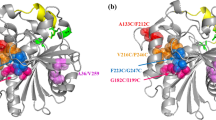
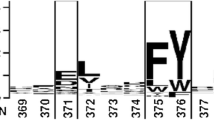
The residues located at the crevice-like binding site of four representative lipases including Thermomyces lanuginosus lipases (TLL and Lip), Rhizopus oryzae lipase (ROL), and Rhizomucor miehei lipase (RML) were identified through structural analysis. The residues at the bottom of the crevice-like binding site recognizing the substrates with short/medium carbon chain length and those located at the right-hand wall of the surface crevice region affecting the product release were changed by site-directed mutagenesis. The corresponding double mutants exhibited ~ 5 to 14-fold higher activity towards p-nitrophenyl esters than their wild types, and their substrate preference shifted to acyl moiety with shorter carbon chain length. In addition, the mutations led to an increase of B-factor, resulting in decrease of their optimum temperature by 10–20 °C.
Conclusions
The key residues located at the crevice-like binding site play important roles in determining lipase activity, substrate preference and optimum temperature, which offers a useful new paradigm for facilitating rational design of lipases.





Similar content being viewed by others
References
Brundiek H, Padhi SK, Kourist R, Evitt A, Bornscheuer UT (2012) Altering the scissile fatty acid binding site of Candida antarctica lipase A by protein engineering for the selective hydrolysis of medium chain fatty acids. Eur J Lipid Sci Technol 114:1148–1153
Ding X, Zheng RC, Tang XL, Zheng YG (2018) Engineering of Talaromyces thermophilus lipase by altering its crevice-like binding site for highly efficient biocatalytic synthesis of chiral intermediate of Pregablin. Bioorg Chem 77:330–338
Durmaz E, Kuyucak S, Sezerman UO (2013) Modifying the catalytic preference of tributyrin in Bacillus thermocatenulatus lipase through in silico modeling of enzyme–substrate complex. Protein Eng Des Sel 26:325–333
Gao C, Lan D, Liu L, Zhang H, Yang B, Wang Y (2014) Site-directed mutagenesis studies of the aromatic residues at the active site of a lipase from Malassezia globosa. Biochimie 102:29–36
Li XJ, Zheng RC, Ma HY, Huang JF, Zheng YG (2014) Key residues responsible for enhancement of catalytic efficiency of Thermomyces lanuginosus lipase Lip revealed by complementary protein engineering strategy. J Biotechnol 188:29–35
Li G, Mariasolano MA, Romerorivera A, Osuna S, Reetz MT (2017) Inducing high activity of a thermophilic enzyme at ambient temperatures by directed evolution. Chem Commun 53:9454
Lorenzo MD, Hidalgo A, Molina R, Hermoso JA, Pirozzi D, Bornscheuer UT (2007) Enhancement of the stability of a prolipase from Rhizopus oryzae toward aldehydes by saturation mutagenesis. Appl Environ Microbiol 73:7291–7299
Marton Z et al (2010) Mutations in the stereospecificity pocket and at the entrance of the active site of Candida antarctica lipase B enhancing enzyme enantioselectivity. J Mol Catal B Enzym 65:11–17
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Pandey A, Benjamin S, Soccol CR, Nigam P, Krieger N, Soccol VT (1999) The realm of microbial lipases in biotechnology. Biotechnol Appl Biochem 29:119–131
Pleiss J, Fischer M, Schmid RD (1998) Anatomy of lipase binding sites: the scissile fatty acid binding site. Chem Phys Lipids 93:67–80
Pleiss J, Fischer M, Peiker M, Thiele C, Schmid RD (2000) Lipase engineering database: understanding and exploiting sequence–structure–function relationships. J Mol Catal B Enzym 10:491–508
Reetz MT (2002) Lipases as practical biocatalysts. Curr Opin Chem Biol 6:145–150
Wang J, Wang D, Wang B, Mei ZH, Liu J, Yu HW (2012) Enhanced activity of Rhizomucor miehei lipase by directed evolution with simultaneous evolution of the propeptide. Appl Microbiol Biotechnol 96:443–450
Yang J, Koga Y, Nakano H, Yamane T (2002) Modifying the chain-length selectivity of the lipase from Burkholderia cepacia KWI-56 through in vitro combinatorial mutagenesis in the substrate-binding site. Protein Eng 15:147–152
Acknowledgements
This work was financially supported by the National High Technology Research and Development Programs of China (No. 2012AA022201), the Key Scientific and Technology Programs of Zhejiang Province (No. 2012C03005-2) and the Natural Science Foundation of Zhejiang Province (No. Z4090612).
Supporting information
Supplementary Table 1—Primers for site-directed mutagenesis of Lip, TLL, ROL and RML.
Supplementary Fig. 1—Sequence alignment of TTL and homologous proteins.
Supplementary Fig. 2—SDS-PAGE analysis of the purified lipases.
Supplementary Fig. 3—Far-UV CD spectra of wild-type lipase and their mutants plotted by scanning 190-400 nm at 35 °C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, X., Tang, XL., Zheng, RC. et al. Identification and engineering of the key residues at the crevice-like binding site of lipases responsible for activity and substrate specificity. Biotechnol Lett 41, 137–146 (2019). https://doi.org/10.1007/s10529-018-2620-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2620-6