Abstract
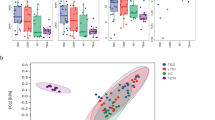
An improved understanding of the lung microbiome may lead to better strategies to diagnose, treat, and prevent pulmonary tuberculosis (PTB). However, the characteristics of the lung microbiomes of patients with TB remain largely undefined. In this study, 163 bronchoalveolar lavage (BAL) samples were collected from 163 sputum-negative suspected PTB patients. Furthermore, 12 paired BAL samples were obtained from 12 Mycobacterium tuberculosis-positive (MTB+) patients before and after negative conversion following a two-month anti-TB treatment. The V3–V4 region of the 16S ribosomal RNA (rRNA) gene was used to characterize the microbial composition of the lungs. The results showed that the prevalence of MTB in the BAL samples was 42.9% (70/163) among the sputum-negative patients. The α-diversity of lung microbiota was significantly less diverse in MTB+ patients compared with Mycobacterium tuberculosis-negative (MTB−) patients. There was a significant difference in β-diversity between MTB+ and MTB− patients. MTB+ patients were enriched with Anoxybacillus, while MTB− patients were enriched with Prevotella, Alloprevotella, Veillonella, and Gemella. There was no significant difference between the Anoxybacillus detection rates of MTB+ and MTB− patients. The paired comparison between the BAL samples from MTB+ patients and their negative conversion showed that BAL negative-conversion microbiota had a higher α-diversity. In conclusion, distinct features of airway microbiota could be identified between samples from patients with and without MTB. Our results imply links between lung microbiota and different clinical groups of active PTB.
Similar content being viewed by others
References
Adami, A.J., and Cervantes, J.L. (2015). The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis 95, 651–658.
Bolger, A.M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.
Botero, L.E., Delgado-Serrano, L., Cepeda, M.L., Bustos, J.R., Anzola, J. M., Del Portillo, P., Robledo, J., and Zambrano, M.M. (2014). Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis. Microbiome 2, 29.
Cheung, M.K., Lam, W.Y., Fung, W.Y.W., Law, P.T.W., Au, C.H., Nong, W., Kam, K.M., Kwan, H.S., and Tsui, S.K.W. (2013). Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS ONE 8, e54574.
Cole, J.R., Wang, Q., Fish, J.A., Chai, B., McGarrell, D.M., Sun, Y., Brown, C.T., Porras-Alfaro, A., Kuske, C.R., and Tiedje, J.M. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucl Acids Res 42, D633–D642.
Cui, Z., Zhou, Y., Li, H., Zhang, Y., Zhang, S., Tang, S., and Guo, X. (2012). Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol 12, 276.
Dheda, K., Barry 3rd, C.E., and Maartens, G. (2016). Tuberculosis. Lancet 387, 1211–1226.
Dickson, R.P., Erb-Downward, J.R., Freeman, C.M., McCloskey, L., Beck, J.M., Huffnagle, G.B., and Curtis, J.L. (2015). Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc 12, 821–830.
Edgar, R.C., Haas, B.J., Clemente, J.C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200.
Geng, J., Fan, H., Tang, X., Zhai, H., and Zhang, Z. (2013). Diversified pattern of the human colorectal cancer microbiome. Gut Pathog 5, 2.
Goh, K.M., Kahar, U.M., Chai, Y.Y., Chong, C.S., Chai, K.P., Ranjani, V., Illias, R.M., and Chan, K.G. (2013). Recent discoveries and applications of Anoxybacillus. Appl Microbiol Biotechnol 97, 1475–1488.
Hong, B.Y., Maulén, N.P., Adami, A.J., Granados, H., Balcells, M.E., and Cervantes, J. (2016). Microbiome changes during tuberculosis and antituberculous therapy. Clin Microbiol Rev 29, 915–926.
Huang, L., Gao, R., Yu, N., Zhu, Y., Ding, Y., and Qin, H. (2019). Dysbiosis of gut microbiota was closely associated with psoriasis. Sci China Life Sci 62, 807–815.
Huffnagle, G.B., Dickson, R.P., and Lukacs, N.W. (2017). The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol 10, 299–306.
Kong, C., Gao, R., Yan, X., Huang, L., He, J., Li, H., You, J., and Qin, H. (2019). Alterations in intestinal microbiota of colorectal cancer patients receiving radical surgery combined with adjuvant CapeOx therapy. Sci China Life Sci 62, 1178–1193.
Krishna, P., Jain, A., and Bisen, P.S. (2016). Microbiome diversity in the sputum of patients with pulmonary tuberculosis. Eur J Clin Microbiol Infect Dis 35, 1205–1210.
Li, F., Sun, G., Wang, Z., Wu, W., Guo, H., Peng, L., Wu, L., Guo, X., and Yang, Y. (2018). Characteristics of fecal microbiota in non-alcoholic fatty liver disease patients. Sci China Life Sci 61, 770–778.
Li, T., Long, M., Ji, C., Shen, Z., Gatesoupe, F.J., Zhang, X., Zhang, Q., Zhang, L., Zhao, Y., Liu, X., et al. (2016). Alterations of the gut microbiome of largemouth bronze gudgeon (Coreius guichenoti) suffering from furunculosis. Sci Rep 6, 30606.
Magoč, T., and Salzberg, S.L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12.
Neff, M. (2003). ATS, CDC, and IDSA update recommendations on the treatment of tuberculosis. Am Fam Physician 68, 1854, 1857–1858, 1861–1852.
Ottesen, A., Ramachandran, P., Reed, E., White, J.R., Hasan, N., Subramanian, P., Ryan, G., Jarvis, K., Grim, C., Daquiqan, N., et al. (2016). Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol 16, 275.
Piewngam, P., Zheng, Y., Nguyen, T.H., Dickey, S.W., Joo, H.S., Villaruz, A.E., Glose, K.A., Fisher, E.L., Hunt, R.L., Li, B., et al. (2018). Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562, 532–537.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F.O. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596.
Schloss, P.D., Westcott, S.L., Ryabin, T., Hall, J.R., Hartmann, M., Hollister, E.B., Lesniewski, R.A., Oakley, B.B., Parks, D.H., Robinson, C.J., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541.
Shao, Y., Arias-Cordero, E., Guo, H., Bartram, S., and Boland, W. (2014). In vivo Pyro-SIP assessing active gut microbiota of the cotton leafworm, Spodoptera littoralis. PLoS ONE 9, e85948.
Sulaiman, I., Wu, B.G., Li, Y., Scott, A.S., Malecha, P., Scaglione, B., Wang, J., Basavaraj, A., Chung, S., Bantis, K., et al. (2018). Evaluation of the airway microbiome in nontuberculous mycobacteria disease. Eur Respir J 52, 1800810.
Teo, S.M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., Holt, B.J., Hales, B.J., Walker, M.L., Hollams, E., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715.
Urbaniak, C., Cummins, J., Brackstone, M., Macklaim, J.M., Gloor, G.B., Baban, C.K., Scott, L., O’Hanlon, D.M., Burton, J.P., Francis, K.P., et al. (2014). Microbiota of human breast tissue. Appl Environ Microbiol 80, 3007–3014.
Wang, Q., Garrity, G.M., Tiedje, J.M., and Cole, J.R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267.
Weinstock, G.M. (2012). Genomic approaches to studying the human microbiota. Nature 489, 250–256.
Wilson, B.A., and Slepecky, R.A. (1962). The lysis of Mycobacterium tuberculosis by microbial enzymes. Am Rev Respir Dis 86, 246–251.
Wipperman, M.F., Fitzgerald, D.W., Juste, M.A.J., Taur, Y., Namasivayam, S., Sher, A., Bean, J.M., Bucci, V., and Glickman, M.S. (2017). Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep 7, 10767.
Wu, J., Liu, W., He, L., Huang, F., Chen, J., Cui, P., Shen, Y., Zhao, J., Wang, W., Zhang, Y., et al. (2013). Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS ONE 8, e83445.
Zhou, Y., Lin, F., Cui, Z., Zhang, X., Hu, C., Shen, T., Chen, C., Zhang, X., and Guo, X. (2015). Correlation between either Cupriavidus or Porphyromonas and primary pulmonary tuberculosis found by analysing the microbiota in patients’ bronchoalveolar lavage fluid. PLoS ONE 10, e0124194.
Acknowledgements
This work was supported by CAMS Innovation Fund for Medical Sciences (2017-I2M-3-017, 2016-I2M-1-013), the 13th-Five-Year National Science and Technology Major Project on the “prevention and treatment of AIDS, viral hepatitis and other infectious diseases” (2018ZX10711001, 2017ZX10201301-002-002), National Natural Science Foundation of China (81525016) and the Non-profit Central Research Institute Fund of CAMS (2019PT31006, 2019PT31007).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Compliance and ethics
The author(s) declare that they have no conflict of interest.
Supporting Information
Methods
Table S1 Demographic and clinical characteristics of the study population
Figure S1 Rarefaction curves showing total observed species per sample across multiple read depths, indicates that 36,000 reads results in good community coverage, demonstrated by curves approaching a plateau.
Figure S2 Bacterial composition of bronchoalveolar lavage fluid (BAL) samples collected from 163 patients and the relative abundance of the most abundant phyla and genera (representing 98.9% of the total reads).
Figure S3 (A–C) Antibiotic administration, age and smoking status showed no difference between microbial community states (MCS). (D) Detection rate of Mycobacterium in BALs varied between microbial community states (MCS) (chi-square test, P=0.006).
Figure S4 Paradigm for evaluating suspected patients with pulmonary tuberculosis.
The supporting information is available online at http://life.scichina.com and https://link.springer.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors.
Online Data Supplemental Text
Rights and permissions
About this article
Cite this article
Hu, Y., Kang, Y., Liu, X. et al. Distinct lung microbial community states in patients with pulmonary tuberculosis. Sci. China Life Sci. 63, 1522–1533 (2020). https://doi.org/10.1007/s11427-019-1614-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-1614-0