Abstract
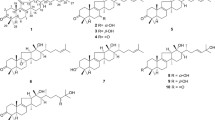
Three new sesquiterpenes, phlebidiol, phlebioic acid, and phlebiolide, as well as the known compound tremetriol, were isolated from cultures of the basidiomycete Phlebia tremellosa. The structures of all isolated compounds were established by extensive spectroscopic analyses, including those involving extensive two-dimensional nuclear magnetic resonance. The absolute configurations of phlebidiol, phlebioic acid, and phlebiolide were determined by comparisons of experimental and calculated electronic circular dichroism spectra. Phlebidiol and phlebioic acid have previously unreported carbon skeletons, for which we propose the skeletal names “seco-sterpurane” and “phlebiane,” respectively. Phlebiolide is also the second published example of a merulane sesquiterpene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Basidiomycetes is a major class of fungi, together with Ascomycetes, and is a rich source of unique secondary metabolites. Phlebia tremellosa (Schrad.) Nakasone & Burds. (formerly Merulius tremellosus), a species in class Basidiomycetes, is classified in the family Meruliaceae and is also known to be one of the wood-decay fungi [1, 2]. Sesquiterpenes with rare skeletons, such as sterpurane, isolactarane, and merulane sesquiterpenes, have been identified in previous studies to be secondary products of P. tremellosa or P. uda [3,4,5]. Although thousands of sesquiterpenes have been identified in the literature, sterpurane and isolactarane sesquiterpenes are rarely found in nature [6]. Following the isolation of the first sterpurane, sterpuric acid, from Chondrostereum purpureum (formerly Stereum purpureum) in 1981 [7], sterpurane sesquiterpenes have been isolated from the basidiomycetes Artomyces pyxidatus (formerly Clavicorona pyxidata) [8], Flammulina velutipesin [9], and Gloeophyllum sp. [10], as well as from Phlebia spp. and the soft coral Alcyonium acaule [11]. Isolactarorufin, the first isolactarane to be identified, was isolated from Lactarius rufus in 1976 [12]. Isolactarane sesquiterpenes have since been isolated from the basidiomycetes Flammulina velutipesin [8] and Phlebia spp., as have sterpurane sesquiterpenes, and have also been detected in Hyphodontia sp. [13], Russula delica [14], and other Lactarius spp. [15]. The only known sesquiterpene with a merulane skeleton is meruliolactone, which was isolated from cultures of Phlebia tremellosa [3].
As part of our research on the secondary metabolites of plant-associated endophytic fungi [16,17,18,19], we isolated and cultured the basidiomycete Phlebia tremellosa from the leaves of Senna alata (Fabaceae) and succeeded in isolating three new sesquiterpenes, namely phlebidiol (1), phlebioic acid (2), and phlebiolide (3), along with a known sterpurane sesquiterpene from solid cultures of P. tremellosa ECN184 (Fig. 1). Compounds 1 and 2 have unprecedented carbon skeletons, for which we propose the skeletal names “seco-sterpurane” and “phlebiane”, respectively. Furthermore, 3 is the second published example of a merulane sesquiterpene.
Results and discussion
Phlebia tremellosa ECN184 was isolated from the healthy leaves of Senna alata and identified by sequencing the D1/D2 26S rRNA gene and internal transcript spacers (ITS) of the ribosomal DNA. The whole mycelia of P. tremellosa, which were cultured on 180 plates of 2% malt extract agar (MEA) for 30 days, were extracted three times with MeOH at room temperature and concentrated under reduced pressure to afford the crude extract. The MeOH extract was then partitioned between ethyl acetate and water. The ethyl acetate extract was subjected to repeated silica gel and Sephadex LH-20 column chromatography (CC) to obtain three new compounds (1–3). In addition, the known compound tremetriol was also isolated and identified based on comparisons of spectroscopic data with spectra published in the literature [3].
Phlebidiol (1), \({[\alpha]{^{20}_{D}}}\) = + 8.4, was isolated as a colorless oil. Its molecular formula (C15H24O3) was established from high-resolution electrospray ionisation mass spectrometry (HRESIMS) data (m/z 275.1607, [M+Na]+, calcd 275.1623). The IR spectrum showed absorptions indicating the presence of hydroxy groups (3402 cm−1) and a carbonyl group (1645 cm−1). The 13C NMR and distortionless enhancement by polarization transfer (DEPT)-135 spectra showed the presence of four methyls, four methylenes, two methines, and five nonprotonated carbons, including three sp2 carbons of a tetra-substituted double bond at δC 159.9 (C-1) and 128.4 (C-9), as well as a ketonic carbon at δC 204.1 (C-8). All protonated carbons were assigned on the basis of the HMQC spectra (Table 1). According to the four degrees of unsaturation and the substructures indicated by the NMR spectra, 1 was proposed to be a bicyclic sesquiterpene. The 1H-NMR data combined with the double-quantum filtered (DQF)–COSY spectrum suggested C-4/C-5/C-6 and C-12/C-13 carbon-chain sequences (Fig. 2). HMBC correlations from one methyl proton signal [δH 1.64 (3H, br s, H3–15)] to C-1, C-8, and C-9 demonstrated the presence of an α-methyl enone moiety, while another methyl resonance [δH 1.16 (3H, s, H3–14)] exhibited four HMBC correlations to two methylene carbons at δC 40.7 (C-6) and 37.8 (C-12), a ketonic carbon (C-8), as well as an undistinguishable carbon signal assigned to a methine carbon at δC 42.8 (C-5) or a quaternary carbon at δC 42.6 (C-7). The observation of four correlations from a methyl-proton singlet indicated a linkage between C-14 and a quaternary carbon, i.e., C-7. Accordingly, the remaining carbon atoms C-6, C-8, and C-12 that correlated with H3-14 were all assumed to be bound to C-7. HMBC correlations from H2-6 [δH 2.19 (1H, dd, J = 4.8, 13.0 Hz); 1.55 (1H, dd, J = 13.0, 14.8 Hz)] to C-8, C-12, and C-14 and from H2-13 [δH 3.73 (2H, m)] to C-7 also sustained the partial structure around C-7. The other methyl proton signals [δH 1.02 (3H, s, H3-10) and δH 1.14 (3H, s, H3-11)] exhibited HMBC correlations with C-2, C-3, C-4, and C-11/C-10. In addition, H2-2 [δH 2.46 and 2.25 (1H each, d, J = 19.4 Hz)] correlated with C-1, C-3, C-4, C-5, and C-9, which was suggestive of linkages from C-1 to C-4 and between C-1 and C-5, as well as the presence of two methyl groups at C-3. Hence, the structure of phlebidiol (1) was proposed to be a four-membered ring-opening product of a sterpurane sesquiterpene. The relative configuration of 1 was confirmed by the essential NOEs [H-2β/H3-10; H3-10/H-5; H-2α/H3-11; H3-11/H-4; H-4/H-6α; H-6α/H3-14] observed in the NOESY spectrum (Fig. 3a).
To elucidate the absolute configuration of 1, the electronic circular dichroism (ECD) spectrum was calculated by quantum-chemical methods and compared with the experimental spectrum. The preliminary conformers, which were generated using the GMMX add-on module (energy window = 10 kcal/mol), were optimized using density functional theory (DFT) at the CAM-B3LYP/6-31+G(d,p) level. ECD spectra of the DFT-optimized conformers were calculated using time-dependent DFT at the CAM-B3LYP/6-31+G(d,p) level with the conductor-like polarizable continuum model (CPCM) solvation model in MeOH. The obtained ECD spectra were Boltzmann averaged on the basis of the calculated relative energies of the DFT-optimized conformers. The experimental and calculated ECD spectra of 1 were in good agreement for the (4R, 5S, 7R) absolute configuration (Fig. 3b).
Phlebioic acid (2), \({[\alpha]{^{20}_{D}}}\) = + 173.8, was obtained as a colorless oil. HRESIMS revealed an [M+Na]+ ion peak at 289.1396, indicating the quasi-molecular formula C15H22O4Na (calcd 289.1416). The IR spectrum exhibited absorptions for hydroxy groups (3435 cm−1) and carbonyl groups (1699 cm−1). The 13C NMR and DEPT135 spectra (Table 1) showed the presence of four methyls, three methylenes, two methines, and six nonprotonated carbons, including four sp2 carbons assigned to a tetra-substituted double bond [δC 132.8 (C-1) and 155.8 (C-10)], a carboxy group [δC 182.8 (C-14)], and a ketonic carbonyl group [δC 208.2 (C-2)]. The DQF–COSY spectrum showed two solitary C-4/C-5/C-6 and C-8/C-9 sequences (Fig. 2). HMBC correlations [H2-8/C-10; H2-9/C-1, C-10, C-15; H3-15/C-1, C-10] indicated that both C-9 and C-15 were bound to an sp2 carbon (C-10) within the double bond. HMBC correlations from H3-13 [δH 1.35 (3H, s)] to two methylene carbons (C-6 and C-8), a quaternary sp3 carbon (C-7), and a carboxy carbon (C-14) suggested the C-6/C-7/C-8 sequence, with both a methyl group and a carboxy group bonded to C-7, as sustained by the HMBC correlations [H2-8/C-6, C-7, C-14 and H2-9/C-7]. Furthermore, both methyl-proton signals [δH 1.00 (3H, s, H3-11) and δH 1.11 (3H, s, H3-12)] correlated with a ketonic carbon (C-2), a quaternary sp3 carbon (C-3), and a methylene carbon (C-4), suggestive of the C-2/C-3/C-4 sequence with two methyl groups at C-3. The HMBC correlation from H2-6 to C-1 confirmed the linkage between C-1 and C-5. The remaining indices of hydrogen deficiency and the chemical shifts of C-1 and C-10 implied a linkage between C-1 and C-2. Hence, the planar structure of 2 was determined to be a sesquiterpene with a previously unreported skeleton. The relative structure of 2 depicted in Fig. 4a was elucidated by the NOEs [H3-11/H-5; H-5/H3-13; H3-12/H-4]. Finally, we determined the absolute structure of 2 to be the (4R, 5S, 7S) configuration because the experimental ECD spectrum was in good agreement with the time-dependent density functional theory (TDDFT) spectrum calculated at the CAM-B3LYP/6-31+G(d,p)//CAM-B3LYP/6-311+G(d,p) level (Fig. 4b).
Phlebiolide (3), \({[\alpha]{^{20}_{D}}}\) = + 21.6, was obtained as a colorless oil. HRESIMS revealed an [M+Na]+ ion at m/z 269.1133, indicating the molecular formula C15H18O3Na (calcd 269.1154). The IR spectrum exhibited a strong absorption for a carbonyl group (1749 cm−1). The 1H NMR spectrum (Table 1) indicated the presence of three methyls, three methylenes, two sp3 methines, and an sp2 methine. Additionally, the 13C NMR spectrum displayed signals for 15 carbon atoms, which could be classified as three methyls, three methylenes, two sp3 and one sp2 methines, and two sp3 and four sp2 nonprotonated carbons, including a carbonyl carbon. The DQF–COSY spectrum revealed correlations indicative of the C-4/C-5/C-6 connection (Fig. 2). The HMBC spectrum showed correlations from both H3-13 and H3-14 to an olefinic methine (C-2), a quaternary carbon (C-3), and a methylene (C-4), as well as mutual correlations (C-14 or C-13). The HMBC correlations [H-2/C-1, C-5; H2-4/C-1, C-2] suggested a linkage between C-1 and C-5, revealing the presence of a 3,3-dimethylcyclopentene moiety. The HMBC spectrum also revealed correlations from the uncoupled methyl group [δH 1.33 (3H, s, H3-15)] to a methine at δC 66.0 (C-6), a nonprotonated carbon at δC 61.0 (C-7), and a methylene at δC 30.5 (C-8). Furthermore, H2-8 correlated with two sp2 quaternary carbons at δC 122.9 (C-9) and 153.8 (C-12) and a carbonyl carbon at δC 174.3 (C-10). The carbon signal of C-12 also correlated with an olefinic proton (H-2) within a 3,3-dimethylcyclopentene moiety. These correlations allowed a seven-membered ring bearing a methyl group and a carbonyl group at C-7 and C-9, respectively, to be constructed. The remaining oxygenated methylene (H2-11) was proposed to be attached to C-12 based on HMBC correlations from H2-11 to C-9 and C-12. The location of C-2 was also corroborated by an NOE between H-2 and H2-11, as observed in the NOESY spectrum. Finally, the correlation from H2-11 to C-10 confirmed the formation of a γ-lactone ring. The oxygen functional groups at C-6 and C-7 were determined to be an epoxy group based on the remaining indices of hydrogen deficiency and the chemical shifts in the 1H and 13C NMR spectra. The presence of the epoxy group was also supported by the fact that the IR spectrum showed no absorption for hydroxy groups. The relative configurations of C-5, C-6, and C-7 were deduced to be as shown in Fig. 5a by key NOEs [H3-13/H-4β; H-4β/H-5; H-5/H-8β; H3-14/H-4α; H-4α/H-6; H3-15/H-8α] in the NOESY spectrum. Based on the comparison of the experimental spectrum and the TDDFT ECD spectrum calculated at the CAM-B3LYP/6-31+G(d,p)//APFD/6-311+G(d,p) level of theory, the absolute structure of 3 was determined to have a (5R, 6S, 7R) configuration (Fig. 5b).
All isolated compounds exhibited no agonistic activities for peroxisome proliferator-activated receptor γ, retinoid X receptor α, and liver X receptor α in a luciferase reportor assay based on the method described in our previous paper [20]. Further studies of the biological activities of isolated compounds are in progress.
In this study, we isolated three new sesquiterpenes with distinct carbon frameworks. Most notably, 1 and 2 have new skeletons that are named “seco-sterpurane” and “phlebiane”, respectively. The biogenetic pathway of phlebidiol (1), the first described seco-sterpurane, is possibly explained by the oxidative aliphatic C–C bond cleavage of the α-hydroxy ketones within the cyclobutane ring of an oxidized tremetriol (Fig. 6). A similar oxidative cleavage of a C–C bond was previously proposed in the clavicoronane sesquiterpenes [7]. Cytochrome P450 17A1 (CYP17A1) is an example of an enzyme that can cleave α-hydroxy ketones devoid of α-C–H bonds. CYP17A1 is a heme enzyme that catalyzes the cleavage of the C-17 to C-20 bond in 17α-hydroxypregnenolone during the biosynthesis of dehydroepiandrosterone via the ferric peroxo-hemiacetal intermediate [21, 22]. Metalloenzymes similar to CYP17A1 seem to be involved even in the biosynthesis of 1. Meanwhile, the first phlebiane sesquiterpene, phlebioic acid (2), is potentially biosynthesized by the secondary ring-opening rearrangement of the isolactarane sesquiterpene, as shown in Fig. 6, although no plausible precursor has been isolated from P. tremellosa. Furthermore, the biosynthetic pathway to phlebiolide (3) also seems to involve the secondary ring-opening rearrangement of an isolactarane sesquiterpene. Merulactone [3], a previously isolated isolactarane from P. tremellosa, is likely to be the precursor of 3; this biogenesis can be rationalized by rearrangement with ring-expansion initiated by the nucleophilic substitution of a hydroxyl group at the C-6 of merulactone. To the best of our knowledge, this is the first example of a proposed scaffold transformation from isolactarane to merulane involving a plausible precursor.
Conclusion
Three novel sesquiterpenes (1–3) were isolated from the cultures of Phlebia tremellosa. The absolute configurations of 1–3 were confirmed by CD spectroscopy and DFT calculations. All new compounds are likely to have originated from sterpurane and/or isolactarane sesquiterpenes. Despite the fact that sterpurane and isolactarane sesquiterpenes have been isolated from several fungal species as mentioned previously, with the exception of merulactone, seco-sterpurane, phlebiane, and merulane sesquiterpenes have never been detected previously [23]. Therefore, our present study revealed the existence of distinctive biosynthetic pathways for the production of rare sesquiterpenes in the basidiomycete P. tremellosa.
Experimental
General experimental procedures
Optical rotation values were recorded on a P-1020 polarimeter (JASCO Corp., Yokyo, Japan). UV spectra were obtained with a Hitachi U-2900 spectrometer (Hitachi Ltd., Tokyo, Japan). ECD spectra were acquired on a JASCO J-820 spectropolarimeter (JASCO Corp.). IR spectra were recorded on a FTIR-8400S spectrophotometer (Shimadzu Ltd., Kyoto, Japan). NMR spectra were acquired on models JNM-AL-400 and JNM-ECZ 400S spectrometers with tetramethylsilane as the internal standard (JEOL Ltd., Tokyo, Japan). Electrospray ionization-MS data were obtained with a LCMS-IT-TOF mass spectrometer (Shimadzu Ltd.). DNA sequencing was performed with a model 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA). Silica gel AP-300 (Toyota Kako, Tokyo, Japan) and Sephadex LH-20 (GE Healthcare, Chicago, IL, USA) resins were used for CC. Silica gel 60 F254 and RP-18 F254S (Merck & Co., Inc., Kenilworth, NJ, USA) were used for TLC.
Fungal material and identification
Phlebia tremellosa ECN184 was isolated from the healthy leaves of Senna alata cultivated in the Herbal Garden of Gifu Pharmaceutical University (Gifu, Japan) in November 2016. The surfaces of the leaves were sterilized by sequential soaking in 95% EtOH for 30 s, 0.5% NaClO for 2 min, and 70% EtOH for 2 min. The surface-sterilized leaves were cut into 1-cm2 pieces and cultured on MEA containing 2% malt extract, 0.1% bacto peptone, 2% d-glucose, and 1.5% agar supplemented with 0.005% chloramphenicol in 9-cm petri dishes. The dishes were then incubated at 27 °C. Emergent organisms were isolated on new MEA. On the basis of the DNA sequencing of the ITS of rDNA and the D1/D2 domain of the 26S rDNA, the isolate belonged to genus Phlebia. The sequence data of P. tremellosa have been deposited at the DNA Data Bank of Japan (DDBJ) under the access numbers LC424440 (26S rRNA) and LC424443 (ITS). The strain was deposited at Department of Microbiology, School of Pharmacy, Aichi Gakuin University (ECN-184).
Fermentation, extraction, and isolation
The fungus P. tremellosa was inoculated onto 150 MEA plates without chloramphenicol. After incubation at 27 °C for 30 days, the fermented materials were extracted with MeOH (2 × 4 L, each 24 h) at room temperature, and the solution was evaporated in vacuo to obtain the MeOH extract (71.8 g). The MeOH extract was partitioned three times with equal amounts of ethyl acetate and water, and the ethyl acetate solution was concentrated under vacuum to yield the ethyl acetate soluble fraction (7.2 g). The ethyl acetate fraction was separated on a silica gel column with CHCl3/MeOH (gradient 50:1 to 8:1, v/v) as the eluent, to give fractions (Frs.) 1–12. Fr. 8 was purified with silica gel CC (n-hexane/ethyl acetate 5:1 to 3:1, v/v) to obtain phlebiolide (3; 2.1 mg). Fr. 9 was subjected to silica gel column purification (n-hexane/ethyl acetate 5:1 to 3:1, v/v) to obtain tremetriol (4.5 mg). Phlebioic acid (2; 2.1 mg) was isolated from Fr. 10 with the aid of a silica gel column (n-hexane/acetone, 1:1). Fr. 11 was purified with a Sephadex LH-20 column (MeOH) to yield phlebidiol (1; 41.4 mg).
Phlebidiol (1)
Colorless oil; \({[\alpha]{^{20}_{D}}}\)+ 8.4 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 246 (3.73), 290sh (2.56); IR (KBr) νmax 3402, 2957, 2932, 2870, 1645, 1459, 1380, 1062, 1041 cm−1; ECD (0.02 mg/mL, MeOH) λext (Δε) 328 (+ 1.20), 247 (–5.24), 203 (+ 2.68) nm; 1H NMR (400 MHz) and 13C NMR (100 MHz) data, see Table 1; HRESIMS m/z 275.1607 [M+Na]+ (calcd for C15H24O3Na, 275.1623).
Phlebioic acid (2)
Colorless oil;\({[\alpha]{^{20}_{D}}}\) + 173.8 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 257 (4.03) nm; IR (KBr) νmax 3435, 2969, 2932, 2874, 1699, 1616, 1464, 1211, 1078 cm−1; ECD (0.02 mg/mL, MeOH) λext (Δε) 346 (+ 2.01), 261 (+ 2.31), 227 (+ 1.42), 217 (+ 1.96), 210 (+ 2.31) nm; 1H NMR (400 MHz) and 13C NMR (100 MHz) data, see Table 1; HRESIMS m/z 289.1396 [M+Na]+ (calcd for C15H22O4Na, 289.1416).
Phlebiolide (3)
Colorless oil; \({[\alpha]{^{20}_{D}}}\) + 21.6 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 275 (4.03) nm; IR (KBr) νmax 2957, 2926, 2864, 1749, 1651, 1647, 1456, 1224, 1159, 1041 cm−1; ECD (0.02 mg/mL, MeOH) λext (Δε) 254 (+ 4.47), 205 (– 5.01) nm; 1H NMR (400 MHz) and 13C NMR (100 MHz) data, see Table 1; HRESIMS m/z 269.1133 [M+Na]+ (calcd for C15H18O3Na, 269.1154).
Computational methods
Conformers of 1–3 were generated using the GMMX add-on module of GaussView 6 with an energy window of 10 kcal/mol. Optimization of suggested conformers followed by TDDFT calculations were performed using Gaussian 16 with various combinations of functionals (B3LYP, CAM-B3LYP, APFD, ωB97X-D) and basis sets [6-311+G(d,p), 6-31+G(d,p)] with the CPCM solvent model. ECD spectra were generated by the SpecDis program using a Gaussian band shape with 0.3–0.35 eV [24, 25]. The overall CD spectra of the obtained conformers were Boltzmann weighted at 298 K after UV correction.
Change history
10 April 2020
The article Sesquiterpenes with new carbon skeletons from the basidiomycete Phlebia tremellosa, written by Ken ichi Nakashima, Junko Tomida, Takao Hirai, Yoshiaki Kawamura and Makoto Inoue was originally published Online First without Open Access.
References
Nakasone KK, Burdsall HH Jr (1984) Merulius, a synonym of Phlebia. Mycotaxon 21:241–246
Schrader HA (1794) Spicilegium florae germanicae. Impensis C. Ritsdheri, Hanover
Sterner O, Anke T, Sheldrick WS, Steglich W (1990) New sterpurane and isolactarane sesquiterpenes from the fungus Merulius tremellosus. Tetrahedron 46:2389–2400. https://doi.org/10.1016/S0040-4020(01)82020-2
Jonassohn M, Anke H, Sterner O, Svensson C (1994) New compounds isolated from the culture filtrate of the fungus Merulius tremellosus. Tetrahedron Lett 35:1593–1596. https://doi.org/10.1016/S0040-4039(00)76767-0
Schuffler A, Wollinsky B, Anke T, Liermann JC, Opatz T (2012) Isolactarane and sterpurane sesquiterpenoids from the basidiomycete Phlebia uda. J Nat Prod 75:1405–1408. https://doi.org/10.1021/np3000552
Fraga BM (2013) Natural sesquiterpenoids. Nat Prod Rep 30:1226–1264. https://doi.org/10.1039/C3NP70047J
Ayer WA, Saeedi-Ghomi MH, Van Engen D, Tagle B, Clardy J (1981) The sterpuric acids: a new type of sesquiterpenoid. Tetrahedron 37:379–385. https://doi.org/10.1016/0040-4020(81)85074-0
Zheng Y, Shen Y (2009) Clavicorolides A and B, sesquiterpenoids from the fermentation products of edible fungus Clavicorona pyxidata. Org Lett 11:109–112. https://doi.org/10.1021/ol8024549
Wang Y, Bao L, Liu D, Yang X, Li S, Gao H, Yao X, Wen H, Liu H (2012) Two new sesquiterpenes and six norsesquiterpenes from the solid culture of the edible mushroom Flammulina velutipes. Tetahedron 68:3012–3018. https://doi.org/10.1016/j.tet.2012.02.021
Rasser F, Anke T, Sterner O (2000) Secondary metabolites from a Gloeophyllum species. Phytochemistry 54:511–516. https://doi.org/10.1016/S0031-9422(00)00137-0
Cimino G, De Giulio A, De Rosa S, De Stefano S (1989) New sterpurane sesquiterpenoid from the mediterranean Alcyonum acaule: structure of 3-acetoxy-sterpurene. Tetrahedron 45:6479–6484. https://doi.org/10.1016/S0040-4020(01)89524-7
Daniewski WM, Kocor M, Thoren S (1976) Constituents of higher fungi. Part VIII. Isolactarorufin, a novel tetracyclic sesquiterpene lactone from Lactarius rufus. Heterocycles 5:77–84
Erkel G, Anke T, Velten R, Gimenez A, Steglich WZ (1994) Hyphodontal, a new antifungal inhibitor of reverse transcriptases from Hyphodontia sp. (Corticiaceae, Basidiomycetes). Naturforsch C 49:561–570
Yaoita Y, Watanabe N, Takano D, Kikuchi M (2004) Sesquiterpenoids from the fruit bodies of Russula delica. Nat Med 58:235
Wang Y, Yang SP, Yue JM, Chow S, Kitching W (2003) Novel sesquiterpenes from the fungus Lactarius piperatus. Helv Chim Acta 86:2424–2433. https://doi.org/10.1002/hlca.200390195
Nakashima K, Tomida J, Hirai T, Morita Y, Kawamura Y, Inoue M (2017) Tubakialactones A–E, new polyketides from the endophytic fungus Tubakia sp. ECN-111. Tetrahedron Lett 58:2248–2251. https://doi.org/10.1016/j.tetlet.2017.04.076
Nakashima K, Tomida J, Kamiya T, Hirai T, Morita Y, Hara H, Kawamura Y, Adachi T, Inoue M (2018) Diaporthols A and B: bioactive diphenyl ether derivatives from an endophytic fungus Diaporthe sp. Tetrahedron Lett 59:1212–1215. https://doi.org/10.1016/j.tetlet.2018.02.032
Nakashima K, Tomida J, Hirai T, Morita Y, Kawamura Y, Inoue M (2017) A new isocoumarin derivative from an endophytic fungus Thielavia sp. isolated from Crassula ovata. Heterocycles 94:117–121. https://doi.org/10.3987/COM-16-13599
Nakashima K, Tomida J, Tabata M, Hirai T, Morita Y, Kawamura Y, Inoue M (2017) A new chromone derivative from an endophytic fungus Xylaria sp. isolated from Ardisia crenata. Heterocycles 94:308–313. https://doi.org/10.3987/COM-16-13625
Nakashima K, Murakami T, Tanabe H, Inoue M (2014) Identification of a naturally occurring retinoid X receptor agonist from Brazilian green propolis. Biochim Biophys Acta 1840:3034–3041. https://doi.org/10.1016/j.bbagen.2014.06.011
Mak PJ, Gregory MC, Denisov IG, Sligar SG, Kincaid JR (2015) Unveiling the crucial intermediates in androgen production. Proc Natl Acad Sci USA 112:15856–15861. https://doi.org/10.1073/pnas.1519376113
Bhattacharya S, Rahaman R, Chatterjee S, Paine TK (2017) Aliphatic C−C bond cleavage in α-hydroxy ketones by a dioxygen-derived nucleophilic iron-oxygen oxidant. Chem Eur J 23:3815–3818. https://doi.org/10.1002/chem.201605672
Quin MB, Flynn CM, Schmidt-Dannert C (2014) Traversing the fungal terpenome. Nat Prod Rep 31:1449–1473. https://doi.org/10.1039/C4NP00075G
Bruhn T, Schaumloffel A, Hemberger Y, Pescitelli G (2017) SpecDis version 1.71. Berlin, Germany. https://specdis-software.jimdo.com. Accessed 7 Jan 2019
Bruhn T, Schaumloffel A, Hemberger Y, Bringmann G (2013) SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 25:243–249. https://doi.org/10.1002/chir.22138
Acknowledgments
This study was suppted by JSPS KAKENHI Grant number JP18K14939 and a research grant from the Institute of Pharmaceutical Life Sciences, Aichi Gakuin University. The authors also thank Dr. Naohito Abe, Prof. Masayoshi Oyama, and Prof. Eiji Sakai at Gifu Pharmaceutical University for the HR-ESIMS measurements and supplying the plant materials.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest associated with this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised due to a retrospective Open Access order.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nakashima, Ki., Tomida, J., Hirai, T. et al. Sesquiterpenes with new carbon skeletons from the basidiomycete Phlebia tremellosa. J Nat Med 73, 480–486 (2019). https://doi.org/10.1007/s11418-019-01286-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11418-019-01286-8