Abstract
Purpose
This study focuses on updating, improving, and expanding the extrapolation factors needed to convert various acute or chronic effect concentration indicators into consistent chronic EC10eq (effect concentration inducing a 10% response over background) for use in life cycle assessment (LCA). Our main objectives include (1) to present a detailed approach for the harmonization of ecotoxicity data, with a focus on deriving extrapolation factors, and (2) to estimate both generic and species group-specific extrapolation factors, facilitating the conversion of effect concentration indicator groups (EC10eq and EC50eq) into chronic EC10eq.
Methods
Experimental ecotoxicity data were sourced from CompTox Version 2.1.1, which integrates toxicity information from ToxValDB v9.1.1, and the information from REACH registration dossiers. We developed a framework for harmonizing ecotoxicity data, ensuring uniformity and high quality of aquatic ecotoxicity information from these sources. Through linear regression analysis, both generic and species group-specific extrapolation factors were then derived.
Results and discussion
Harmonization of ecotoxicity data yielded a streamlined dataset with 339,729 datapoints for 10,668 chemicals, reflecting a 54% reduction in raw datapoints. The geometric mean-based aggregation process produced 79,001 aggregated effect concentration datapoints at the species level, 41,303 at the species group level, and 23,215 at the effect concentration indicator level for these chemicals. This process facilitated the derivation of 3 generic and 24 species group-specific extrapolation factors, allowing for the conversion of effect concentration indicator groups (EC10eq and EC50eq) to a chronic EC10eq across two exposure classes (acute vs. chronic) and species groups, as defined in the US EPA ECOTOX knowledgebase, including algae, amphibians, fish, crustaceans, insects/spiders, invertebrates, molluscs, and worms.
Conclusions
The harmonization of ecotoxicity data and the derived extrapolation factors permit the integration of diverse datapoints with varying effect concentration indicators and exposure durations into USEtox ecotoxicity characterization factors. This has the potential to enhance substance coverage for characterizing ecotoxicity effects across chemicals in LCA frameworks by permitting wider species coverage. More generally, this is part of global efforts to extend the potential for quantitative assessment of environmental impacts of chemicals in an LCA framework.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Life cycle considerations are increasingly part of chemical assessment frameworks to evaluate the potential toxicity impacts of chemicals in a product or service (Tickner et al. 2021) using either life cycle thinking or more detailed life cycle assessment (LCA) (Jacobs et al. 2016). LCA methods are constantly evolving, but according to its International Organization for Standardization (ISO) definition, an analyst performing an LCA must perform a life cycle impact assessment (LCIA—the “phase of LCA aimed at understanding and evaluation the magnitude and significance of the potential environmental impacts”) which typically demands “characterization factors” (CFs) that convert resource consumption and pollutant emissions to potential impacts (ISO 2006). For freshwater ecotoxicity impacts, fate-exposure-effect models such as USEtox can be employed to calculate CFs (Fantke et al. 2017; Hauschild et al. 2008; Rosenbaum et al. 2008). USEtox is the UNEP/SETAC scientific consensus model for human toxicity and ecotoxicity CFs (Fantke et al. 2017). In the USEtox framework, freshwater ecotoxicity CFs (PAF m3 day/kg-emitted) are computed by integrating three specific factors in a matrix system: fate factors (FF, kg kg−1 day−1), environmental exposure factors (XF), and freshwater ecotoxicological effect factors (EF, PAF m3 kg−1) (Fantke et al. 2017; Owsianiak et al. 2023). This relationship is succinctly captured by the equation: CF = FF × XF × EF (Fantke et al. 2018). USEtox framework is the basis of current European Product Environmental Footprint (PEF) norms (EU 2023).
Analysts suffer from a critical shortage of CFs. This shortage is apparent if we compare the pre-registered substances database of the European Chemicals Agency (ECHA), which contains 145,299 unique substances/entries (ECHA 2023), to the USEtox database (version 2.01), which has only 3077 organic substances and 27 inorganic substances (Fantke et al. 2017). The challenge of managing the shortage of CFs today primarily involves generating more extensive experimental laboratory toxicity data including using in vitro methods along with finding ways to extrapolate from existing data to suit LCA purposes (Chang et al. 2022; Zhang et al. 2018). The extension of experimental data includes both in vitro and in vivo data. However, due to the difficulty in extrapolating from in vitro to in vivo effects (Rodea-Palomares & Bone 2024), this paper only uses in vivo data to generate extrapolation factors.
New experimental toxicity data is becoming increasingly available as laboratory testing and digitalization of data proceeds, which offers data availability to calculate CFs for more chemicals (Kristiansson et al. 2021). At present, the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) dossiers and CompTox stand out as some of the most extensive and up-to-date online repositories for ecotoxicity information on chemicals, recognized as valuable data sources in numerous regulatory and academic domains (Fantke et al. 2020; Gustavsson et al. 2017; Lowe & Williams 2021; Müller et al. 2017; Saouter, Biganzoli, et al., 2019; Saouter, Wolff, et al. 2019; Silva 2020; Williams et al. 2021). CompTox is the US Environmental Protection Agency (EPA) web-based chemistry dashboard that integrates chemical data, including physicochemical properties, environmental fate and transport, exposure, usage, in vivo toxicity, and in vitro bioassay data (Williams et al. 2017). Nevertheless, the generation of robust CFs necessitates careful data curation and harmonization that must consider, among other aspects, the test species, the units in which results are expressed, different effect concentration indicators, and exposure durations.
A new method for calculating USEtox EFs was proposed by Owsianiak et al. (2023) based on the collaborative efforts of the Ecotoxicity Task Force and the Society of Environmental Toxicology and Chemistry (SETAC) Pellston workshop. Whereas CFs have historically been based on chronic EC50 values, one of the new recommendations is an HC20-based approach. HC20 is the hazardous concentration affecting 20% of species in an exposed ecosystem, identified on an SSD curve of chronic EC10-equivalent ecotoxicity data (Sala et al. 2022). The chronic EC10 data recommended by Owsianiak et al. (2023) for calculating HC20 are relatively scarce as illustrated by Douziech et al. (2024) and Saouter et al. (2018). To fill the data gaps, extrapolation factors are often used to extrapolate ecotoxicity data from different effect concentration indicators with different exposure durations and species groups to chronic EC10 equivalents.
Extrapolation factors that convert between effect concentration indicators like acute and chronic NOEC (no‐observed effect concentration), EC50 (concentration inducing a 50% response), and EC10 (concentration inducing a 10% response) values are used to fill data gaps in CF calculation (Aurisano et al. 2019). Current extrapolation methods either employ predefined assessment factors (ECHA 2012) or utilize generic acute-to-chronic extrapolation ratios derived from a limited set of chemicals (Fantke et al. 2017; Payet 2004). These methods consider the type of effect concentration indicators and exposure durations but often overlook variations across species groups, which can be significant as illustrated by Aurisano et al. (2019). Several studies have examined the provision of extrapolation factors in aquatic environment, both at generic and species group-specific levels. Notable among these are works by Aurisano et al. (2019), Payet (2004), De Zwart (2002), and Saouter, Wolff, et al. (2019). Specifically, Aurisano et al. (2019) analyzed ECHA 2018 data, comprising 71,343 raw datapoints for 1927 chemicals. Their harmonized dataset, which distilled down to 9627 datapoints for 1048 chemicals, offered extrapolation factors for three species groups, algae/cyanobacteria, crustaceans, and fish, spanning three effect concentration indicators: EC50, NOEC, and EC10. Payet (2004), drawing from sources like ECETOC (European Centre for Ecotoxicology and Toxicology of Chemicals) and US EPA, presented data on 134,088 datapoints across three species groups, plants and algae, vertebrates, and invertebrates, covering effect concentration indicators EC50, LOEC, and NOEC. While Payet (2004) provided extrapolation factors to convert varied exposures and effect concentration indicators into EC50, the factors were not species group-specific, and the data harmonization approach was not fully explained. Saouter, Wolff, et al. (2019) utilized ECHA 2015 data, starting with 305,068 raw datapoints, refining them to a harmonized set of 54,353. They provided extrapolation factors for three species groups, algae, crustaceans, and fish, focusing on effect concentration indicators EC50 chronic, EC50 acute, and NOEC chronic. Currently, for extrapolation factors, the literature only covers a limited range of species groups, and the endpoints EC50, NOEC, LOEC, and EC10 vary. This highlights an opportunity for further research, particularly regarding the development of additional species group-specific extrapolation factors for other species groups and harmonizing the classification of effect concentration indicators into two categories: EC10eq and EC50eq. Additionally, the inclusion of more data can expand the number of chemicals that the extrapolation factors are based on, thereby improving their representativeness and accuracy.
The primary ambition of this study is to improve the use of available ecotoxicity data by generating new extrapolation factors to enable the conversion of effect concentration indicators (EC10eq and EC50eq) to a chronic EC10eq, based on experimental data. Our approach is twofold: firstly, we present a strategy for harmonizing ecotoxicity data, which will aid in determining extrapolation factors. Secondly, we introduce both generic and species group-specific extrapolation factors, tailored to diverse effect concentration indicators and exposure durations. These proposed factors are specifically designed for their application in the ecotoxicity characterization factor computation within the latest LCA framework. We also present factors for effect concentration conversion to chronic EC50eq values in the supplementary data to allow compatibility with the current PEF norms.
2 Methods
2.1 Ecotoxicity data collection
To generate the extrapolation factors, aquatic ecotoxicity data were gathered from REACH dossiers and CompTox (Adkins 2023; REACH 2020; Williams et al. 2017). The REACH data was accessed in August 2020, while CompTox data was procured in July 2023.
In the REACH database, dossiers are systematically organized into sections, with subsections detailing the outcomes of various studies. For the purposes of this research, which centers on aquatic ecosystems, the “ecotoxicological information” section was chosen. Within this, the “aquatic toxicity” subsection was of primary interest. This subsection encompasses a range of studies, including but not limited to short- and long-term toxicity evaluations for fish and aquatic invertebrates, assessments of toxicity to aquatic algae, cyanobacteria, and other aquatic plants, as well as toxicity tests for microorganisms. Additionally, it covers endocrine disrupter testing in aquatic vertebrates conducted in vivo and toxicity studies for other aquatic entities. Studies within this framework provide the specific effect concentration indicator supplemented with pertinent details such as testing methodology, conditions under which tests were conducted, organisms tested, and the duration of the test (REACH 2020). The accessed database contained 225,517 records providing toxicity information on 12,411 chemicals with an EC Number.
The CompTox data was sourced directly from the US EPA ToxValDB (version 9.4), a comprehensive experimental toxicity database assembled from 49 public sources, with REACH dossier data being an exception. Of these sources, ten specifically offer ecotoxicity data, namely COSMOS (Experimental data for cosmetics ingredients compiled by the COSMOS consortium, https://www.ng.cosmosdb.eu/), DOE ECORISK (Ecological screening data compiled by Los Alamos National Laboratory, https://rais.ornl.gov/documents/ECO_BENCH_LANL.pdf), DOE Wildlife Benchmarks (Wildlife benchmark data compiled by Oak Ridge National Laboratory, https://rais.ornl.gov/documents/tm86r3.pdf), ECHA IUCLID (REACH dossier studies, https://echa.europa.eu/information-on-chemicals/registered-substances), ECOTOX (Data for ecological species, https://cfpub.epa.gov/ecotox/), EFSA (Data from OpenFoodTox, compiled from The European Food Safety Authority (EFSA) risk assessment documents, https://www.efsa.europa.eu/en/data-report/chemical-hazards-database-openfoodtox), EnviroTox_v2 (Data from the Envirotox database for ecological species, https://envirotoxdatabase.org/), HAWC Project (Toxicology study data extracted into HAWC for several public projects, https://www.epa.gov/risk/health-assessment-workspace-collaborative-hawc), HEAST (Provisional risk assessment values from the EPA Health Effects Assessment Summary Tables (HEAST), https://cfpub.epa.gov/ncea/risk/recordisplay.cfm?deid=2877), and HPVIS (Experimental toxicity data from the EPA High Production Volume Information System (HPVIS), https://chemview.epa.gov/chemview/). The latest iteration of ToxValDB can be accessed via the EPA, CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/) (Judson 2018; USEPA 2023). This study contains 517,067 datapoints, detailing ecotoxicity information for 8640 chemicals from CompTox, though it is noteworthy that this data may include duplicate records.
2.2 Selection and harmonization framework
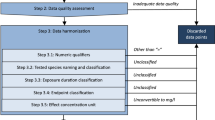
We devised a framework for data selection and harmonization building on the previous work by Aurisano et al. (2019), Aurisano et al. (2023), Saouter, Wolff, et al. (2019), and Aggarwal et al. (2024) to ensure a high-quality dataset for extrapolation across effect concentration indicators, exposure durations, and test species group. Chemical substances with unsatisfactory or missing information, as defined in the data harmonization steps, were excluded. The data was curated according to five main steps as shown in Fig. 1 including chemical identification, data reliability control, data harmonization, consistency checking, and selection of ecologically relevant effects.
The data curation begins with chemical identification. Thus, chemicals lacking Chemical Abstracts Service (CAS) registry numbers were excluded to ensure that each datapoint is accurately linked with its corresponding chemical. Following this, the data reliability control step ensures the integrity of the data by using metadata to filter out information of lesser reliability. For example, ECHA data with Klimisch scores 3 and 4 are excluded (Klimisch et al. 1997). During the data harmonization step, uniformity is achieved in species naming and grouping based on the nomenclature found in the US EPA ECOTOX knowledgebase (U.S.EPA 2023). Effect concentration indicators are classified into two categories: EC10eq and EC50eq. Study durations are harmonized into days, and exposure duration classes are assigned into two categories: acute and chronic. Additionally, concentration units are converted to milligrams per liter (mg/L). The subsequent consistency check verifies that all gathered information is purely experimental, is free from data gaps, and does not include duplicates. The data is collected from different sources, so duplicate records may exist, making duplicate exclusion important. Finally, the relevance of effects at the aquatic ecosystem level was assessed to exclude datapoints with ambiguous relevance to aquatic ecotoxicity, such as biomarkers encompassing biochemistry and genetics. The data is processed using Microsoft Excel. Detailed descriptions of these steps are available in the Supplementary Information in Sect. 1.1.
One of the most important harmonization steps, distinct from previous studies by Aurisano et al. (2019), Payet (2004), De Zwart (2002), and Saouter, Wolff, et al. (2019), is related to the harmonization of effect concentration indicators and their grouping into two categories: EC10eq and EC50eq. In previous studies, various classifications of effect concentration indicators were used, including EC50, NOEC, LOEC, and EC10. For example, Aurisano et al. (2019) combined LOEC and EC10, while Saouter, Wolff, et al. (2019) included NOEC, LOEC, and EC10 to EC20 under the NOECeq category, as well as the threshold of toxicological concern (TTC). The change is primarily because it has long been recognized that the biological effects observed at concentrations reported as NOECs typically affect between 10 and 30% of the exposed populations in the tests (Crane & Newman 2000; Moore & Caux 1997; US EPA 1991). This has led to a long debate on the usability of NOEC concentrations, and direct inferences of specific low-effect concentrations (e.g., the EC10) have been suggested as suitable alternatives (Azimonti & et al. 2015). Currently, NOEC and EC10 are used interchangeably in the derivation of safe concentrations within many regulatory frameworks, such as the European Union REACH regulation, the plant protection product regulation, and the Water Framework Directive (ECHA 2008; EFSA 2013; European Commission 2011). Therefore, in this study, the 10% effect concentration equivalent (EC10eq) group comprises effect concentration indicators EC0, EL0, IC0, LC0, NOAEC, NOEC, NOEL, EC1 to EC10, IC10, LC10, and LOEC. The 50% effect concentration equivalent (EC50eq) group encompasses effect concentration indicators reported as EC50, ErC50, ErL50, IC50, and LC50. Datapoints that could not be classified into these two groups or had missing information were excluded. Additional steps incorporated into this study, which expand upon the previous work by Aurisano et al. (2019) and Saouter, Wolff, et al. (2019), include the use of numeric qualifiers (Step 3.1), consistency checking to remove duplicates (Step 4), and a focus on ecologically relevant effects only (Step 5). In this study, potential duplicates were identified based on similarities in the metadata. For the REACH data, potential duplicates were identified based on having the same CAS, type of information, reliability, media type, effect, numeric qualifier, common species name, species group, effect concentration indicator, and effect concentration units and value. For CompTox, duplicates were identified based on the same CAS, media type, critical effect, year of the data, data reference source, reference title, author of the data, numeric qualifier, common species name, species group, effect concentration indicator, and effect concentration unit and value. It should be noted that distinguishing between freshwater and marine environments in the aquatic system could lead to a higher level of harmonization; however, at least for pesticides, there is no evidence for any systematic differences in sensitivity between these environmental compartments (Klok et al. 2012). Based on toxicity data in the database of De Zwart (De Zwart 2002) and added toxicity data from recent publications, we concluded that there is no systematic difference in toxicity between fresh and saltwater species (including marine and estuarine species).
In this study, we also attempted to provide generic extrapolation factors for available chemicals based on a classification according to the EPA DSSTox Tree as provided by PubChem Classification Browser (https://pubchem.ncbi.nlm.nih.gov/classification/#hid=105). In total, 15 chemical groups were selected as detailed in the supplementary information in Table G2.
2.3 Ecotoxicity extrapolation factor calculation strategy
After harmonization, the primary attributes retained from both databases for subsequent analysis were the Chemical Abstracts Service (CAS) registry number, tested species naming and grouping, exposure classification, effect concentration indicator classification, and effect concentration units.
The refined and harmonized data provides a curated dataset for each chemical. Every datapoint is categorized by a unique species name, species group, exposure class, and effect concentration indicator classification. The calculations occur at three levels: initially at the species level, followed by the species group level, and finally at an overarching generic (species group independent) level as shown in Table 1. For the species level, the geometric mean was calculated for each combination of specific chemical, exposure class, effect concentration indicator classification, species group, and species name. This approach produces an aggregated effect concentration for each chemical, defined by its effect concentration indicator, exposure class, species group, and species name. At the species group level, the geometric mean is derived from the species-level data associated with each particular chemical, exposure class, effect concentration indicator, and species group, such as melamine (CAS: 108–78-1) chronic EC10eq Algae or melamine acute EC10eq Algae. Lastly, for the generic level, the geometric mean is determined using data from the species group level, linked to a specific chemical, exposure class, and effect concentration indicator classification, thus leading to an overall effect concentration for each chemical based on its effect concentration indicator and exposure class.
The harmonized dataset served as the foundation for deriving extrapolation factors. These factors were determined through pairwise comparisons of effect concentration indicator combinations (EC10eq and EC50eq) for chemicals with data spanning across both exposure classes (acute vs. chronic) and various species groups, including algae, amphibians, fish, crustaceans, aquatic plants (including flowers, trees, shrubs, and ferns), fungi, insects/spiders, invertebrates, molluscs, moss and hornworts, and worms. All effect concentration data underwent log10 transformation before comparisons. Regression analysis assessed the correlation between paired effect concentration indicators. Initially, generic extrapolation factors were proposed for effect concentration indicator combinations across exposure classes, irrespective of species groups, by merging data from all species groups. Subsequently, species group-specific extrapolation factors were developed for effect concentration indicator combinations across different exposure classes and species groups. Note that in this study, the extrapolation factor is conceptualized as a multiplier. When applied to the effect concentration indicator under examination, it converts the value to the desired extrapolated chronic EC10eq measurement.
In our calculations, we employed linear regressions in two distinct manners: one with a free slope and the other with a slope set to 1. In both cases, the purpose of these regressions was to gauge the strength of correlation, as denoted by the coefficients of determination (R2), and to estimate the extrapolation factors. Specifically, the free-slope regression was utilized to derive the regression equation for converting various effect concentration indicators to chronic EC10eq across different exposure classes and species groups. On the other hand, the regression with a slope set to unity was instrumental in determining the default extrapolation factors for the same conversion as used by previous studies including Aurisano et al. (2019). Through pairwise comparison, we derived a regression equation with a free slope in the form: log(effect concentration indicator1) = a + b*log(effect concentration indicator2). This translates to effect concentration indicator1 = effect concentration indicator2b × 10a. When the slope is constrained to unity (b = 1), the equation simplifies to effect concentration indicator1 = effect concentration indicator2 × 10a, where 10a represents the default linear extrapolation factor.
3 Results and discussion
3.1 Selection and harmonization framework results
The data retrieved from REACH and CompTox were used as raw inputs in the framework for data selection and harmonization, ensuring a high-quality dataset for extrapolation. Initially, the ECHA database provided toxicity information on 12,411 chemicals with EC Numbers, encompassing 225,517 records. In contrast, the CompTox database contained 517,067 datapoints detailing ecotoxicity information for 8640 chemicals. After the harmonization process (Fig. 1 and Sect. 2.2 in brief, supplementary information in detail), the harmonized data after Step 5 comprised a streamlined dataset of 339,729 datapoints for 10,668 chemicals (Table 2).
3.2 Harmonized data aggregation results
Each datapoint in the dataset is uniquely categorized by its CAS, species name, species group, exposure duration class, and effect concentration indicator classification. The refined and harmonized data were then aggregated at three distinct levels: initially at the species level, then at the species group level, and finally at the effect concentration indicator level. For the species level, the geometric mean of the datapoints—categorized by a unique CAS, species group, exposure class, and effect concentration indicator—was calculated, yielding 79,001 aggregated effect concentration datapoints for 10,668 chemicals. For the species group level, the geometric mean was derived from the species-level data associated with a specific chemical, exposure class, effect concentration indicator, and species group. This process resulted in 41,303 aggregated effect concentration datapoints for the same set of chemicals. Lastly, at the effect concentration indicator level, the geometric mean was determined using data from the species group level associated with a particular chemical, exposure class, and effect concentration indicator classification. This aggregation produced 23,215 effect concentration datapoints for the 10,668 chemicals.
3.3 Ecotoxicity extrapolation factor results
The final harmonized dataset, once aggregated, served as the foundation for deriving a consistent set of extrapolation factors. Before making comparisons, all effect concentration data underwent a log10 transformation. If fewer than 50 overlapping chemicals were available for any pairs, the regression was not performed. If there were multiple datapoints available, aggregation was done using the geometric mean, as detailed in Sect. 3.2. Pairwise correlations were then conducted to evaluate the linear correlation between paired effect concentration indicators, with the correlation coefficient, r, gauging the strength and direction of the linear relationship between these effect concentration indicators. Subsequently, linear regressions, both with a free slope and with a slope forced to unity, were executed to assess the correlation strength, as indicated by the coefficients of determination (R2). These regressions were performed between paired effect concentration indicators to compute generic extrapolation factors and then within each species group to determine species group-specific extrapolation factors.
The primary objective of this study was to identify extrapolation factors that can convert various effect concentration indicators to EC10eq chronic, in line with the requirements of USEtox recommendations. This conversion is essential to effectively utilize the available ecotoxicological effect data in the calculation of characterization factors through extrapolation factor computation. Consequently, extrapolation factors were first calculated for each species group as shown in Table 3 along with Fig. 2 and subsequently generic default extrapolation factors as shown in Table 4 along with Fig. 3 to transform different effect concentration indicators at varying exposure levels to EC10eq chronic. Additionally, the study also provides the best-fit regression equation specifically for EC10eq chronic conversion.
Pairwise comparison of various ecotoxicological effect concentration indicators (log mg/L), as detailed in Table 3. The red line depicts a linear regression fit with an unconstrained slope, while the green line showcases a linear regression fit where the slope is set to unity. The dotted line represents the 95% confidence interval of the linear regression fit with an unconstrained slope (n = number of chemicals, r = correlation, and R2 = Rsquare)
Pairwise comparison of various ecotoxicological effect concentration indicators (log mg/L), as detailed in Table 4. The red line depicts a linear regression fit with an unconstrained slope, while the green line showcases a linear regression fit where the slope is set to unity. The dotted line represents the 95% confidence interval of the linear regression fit with an unconstrained slope (n = number of chemicals, r = correlation, and R2 = Rsquare)
Table 3 and Table 4 present both generic and species-specific extrapolation factors (with the slope set to unity) alongside the available datapoints for each regression. For several combinations, particularly for species groups of aquatic plants, fungi, moss hornworts, and reptiles, data were insufficient to support robust regressions, so extrapolation factors were not calculated. Consequently, for these particular species groups, one might consider applying generic default extrapolation factors at the effect concentration indicator level or resorting to extrapolation factors from closely related species groups, if available.
The species group-specific extrapolation factors exhibit considerable variation across different species groups, as shown in Table 3. In many cases, these values differ considerably from the overall generic extrapolation factors. These differences might arise from the diverse data availability across species groups and effect concentration indicators, with data predominantly coming from algae, crustaceans, and fish. Such findings suggest that studies relying solely on generic extrapolation factors, while overlooking more detailed data for specific species groups, could introduce significant uncertainties. The magnitude of this uncertainty may vary depending on the species group, effect concentration indicator, exposure duration, and the number of datapoints considered. For example, converting acute EC10eq data to chronic EC10eq for algae using a generic default extrapolation factor of 0.3 instead of a species group-specific default extrapolation factor of 0.79 would underestimate extrapolated chronic EC10eq by a factor of 0.79/0.3 = 2.63.
As illustrated in Table 3 and Table 4, robust correlations with R2 > 0.80 were not observed when comparing EC10eq chronic from any effect concentration indicator with either acute or chronic exposure. However, correlations were weaker when acute effect concentration indicators were compared to EC10eq chronic than when chronic datapoints were compared with the same effect concentration indicators within a species group. This highlights that comparisons within the same exposure type across a combination of effect concentration indicators and species groups tend to be more consistent than those across different exposure types.
Given that default ecotoxicity extrapolation factors were derived using a simplified regression approach with a slope set to unity, while regression equations were based on best-fit regressions with a free slope, it is essential to approach the use of these factors judiciously. When studies necessitate EC10eq chronic values, the general approach should be to source unextrapolated EC10 chronic values, whether through direct experimentation, literature reviews, in silico tools, or other relevant methods (Fantke et al. 2017). If experimental EC10 chronic values are not readily available, then extrapolation factors can be employed, but with caution. Best-fit regressions with a free slope (i.e., two parameters) are generally preferred over default (single parameter) ecotoxicity extrapolations, both generically and at the species group level. When considering specific species, it is advisable to prioritize species group-specific extrapolation factors over generic ones, provided that relevant species group information is available. In situations where species group-specific factors are not available for certain species groups, one could either apply the generic default extrapolation factors or use their own judgement to select a species group related to the species in question. Additionally, given that correlations vary in strength across species groups, effect concentration indicators, and exposure durations as depicted in Table 3 and Table 4, chronic extrapolations are preferred over acute ones, especially for species groups with higher correlations.
The REACH and CompTox data underwent rigorous harmonization using a framework, ensuring a high-quality dataset for the extrapolation factor calculation strategy rooted in regression analysis; the process was stringent, leading to a 51% reduction in datapoints. Nevertheless, we were challenged by limitations and gaps in available information. For instance, databases fail to implement checks to update old species names when changes are made leading to uncertainty in species naming and classification. Additionally, exposure classifications present inconsistencies. In the case of algae, tests do not distinguish between acute and chronic effects, which is why we adopted a 24-h exposure duration threshold (Aurisano et al. 2019). Given the rapid reproduction rate of algae, these tests inherently lean towards chronic evaluation (Hahn et al. 2014). Effect concentration indicator classification remains ambiguous due to significant uncertainties, especially in the lower range of species sensitivity distributions, making it challenging statistically to distinguish between NOEC, LOEC, and EC 1–10 values (Iwasaki et al. 2015). One of the most significant challenges faced during the harmonization process is neglecting the effect type. For a given effect concentration indicator, species, and exposure type, tests can analyze a broad spectrum of effects. The sensitivity of these effects can cause variations in exposure concentrations, leading to inconsistencies that are challenging to harmonize given data constraints. These factors can significantly influence the outcomes of ecotoxicity tests and, by extension, our derived extrapolation factors.
In addition, individual datapoints and consequently the pairs used to derive the extrapolation factors can be influenced by several additional factors, for example, the merger of several similar effect concentration indicators into EC10eq (see Sect. 2.2). However, as discussed in the method, the NOEC is itself not indicative of any specific level of effect. Likewise, simply classifying species based on the grouping used by the US EPA ECOTOX knowledgebase (U.S.EPA 2023) might miss relevant relationships which could be observed using a classification of higher taxonomic resolution. Utilizing detailed taxonomic information, such as more relevant taxonomy grouping, species name, genus, family, order, and phylum for each species, as identified by Saouter et al. (2018), can enhance the analysis and drive the development of a comprehensive database of species with detailed taxonomic information to be used in relation to toxicological data. There is also a possibility that individual chemical groups behave differently and that extrapolation factors should be calculated by group. Finally, the way the average is calculated for groups of datapoints, e.g., using the arithmetic mean or the geometric mean, could influence the final results. To assess these uncertainties, we also calculated for specific species groups and generic extrapolation factors, as detailed in the supplementary information from Table S1 to Table S12. The default generic extrapolation factors for different scenarios to EC50eq chronic are within the range of 0.48 to 1.07 from EC10eq acute, 2.47 to 4.90 from EC10eq chronic, and 0.19 to 0.45 from EC50eq acute, with median values of 0.88, 3.17, and 0.36, respectively. These correspond to values of 0.97, 3.38, and 0.45 calculated for all chemicals in this study. For extrapolation to EC10eq chronic, the factors range from 0.15 to 0.31 from EC10eq acute, 0.06 to 0.14 from EC50eq acute, and 0.20 to 0.40 from EC50eq chronic, with median values of 0.30, 0.13, and 0.32. These correspond to values of 0.30, 0.14, and 0.30 calculated for all chemicals in this study. The largest differences were observed when using arithmetic mean aggregation, which resulted in lower R2 values along with higher uncertainty, particularly in the marine ecosystem due to the lack of representative data compared to the freshwater ecosystem. These variations are more pronounced in species-specific extrapolation factors, which we therefore consider the next natural step to investigate in order to provide further refinement of extrapolation factors.
3.4 Comparison of ecotoxicity extrapolation factors with prior research
When examining extrapolation at the species level, there are differences between the factors calculated in this study and those from previous research as shown in Table 5. It should be noted that effect concentration indicators vary across different studies and that here EC10eq and EC50eq, which involve combining different effect concentration indicators, as detailed in Sect. 2.2. However, other studies may use some of these indicators independently or in different combinations. For example, Aurisano et al. (2019) combined LOEC and EC10, while Saouter, Wolff, et al. (2019) included NOEC, LOEC, and EC10 to EC20 under the NOECeq category, as well as the threshold of toxicological concern (TTC).
For the transition from EC50eq acute to EC10eq chronic, Aurisano et al. (2019) determined species group-specific factors of 0.13 (n = 26) and 0.30 (n = 93) for fish and invertebrates, respectively. For the transition from EC50 acute to NOEC chronic, Aurisano et al. (2019) determined factors of 0.16 (n = 54), 0.18 (n = 155), and 0.49 (n = 11) for fish, invertebrates, and algae respectively. In contrast, Saouter, Wolff, et al. (2019) found factors of 0.09 (n = 322), 0.09 (n = 876), and 0.24 (n = 2342) for fish, crustaceans, and algae. Our study presents different values: 0.07 for fish, 0.05 for invertebrates, 0.12 for algae, and 0.09 for crustaceans. For the extrapolation from EC50eq chronic to EC10eq chronic, Aurisano et al. (2019) reported factors of 0.65 (n = 9) for fish, 0.52 (n = 76) for invertebrates, and 0.45 (n = 312) for algae and for the transition from EC50eq chronic to NOEC chronic, factors of 0.29 (n = 19) for fish, 0.37 (n = 89) for invertebrates, and 0.26 (n = 399) for algae. Our study however used EC10eq combining NOEC and EC10 as described in Sect. 2.2 in brief and in the supplementary information in detail, the factors found were 0.25 for fish, 0.36 for invertebrates, and 0.28 for algae. Moving from NOEC acute to EC10eq chronic, Aurisano et al. (2019) identified factors of 0.25 (n = 12) for fish and 0.65 (n = 77) for invertebrates, whereas our study determined them to be 0.2 and 0.42 respectively. The species group-specific extrapolation factors calculated in this study, compared to those reported in previous literature, differ by a maximum factor of 4 for algae, 2.6 for crustaceans, 2.6 for fish, and 6 for invertebrates (a very heterogenous grouping).
Comparing the default generic extrapolation factors from this study with those previously published offers insights into the evolution of the results as more data is incorporated. For instance, when extrapolating from EC50eq acute to EC10eq chronic, our study determined a default generic extrapolation factor of 0.14. This differs from the 0.25 and 0.20 found in studies by Aurisano et al. (2019) for EC10eq chronic and NOEC chronic respectively; however, it roughly aligns with the results from Länge et al. (1998), Warne et al. (2015), King et al. (2017), and Posthuma et al. (2019). In the case of extrapolating from EC50eq chronic to EC10 eq chronic, our derived factor is 0.3, which is different from 0.21 proposed by Payet (2004) for NOEC chronic and 0.5 by Aurisano et al. (2019) for EC10eq chronic but similar to 0.3 proposed by Aurisano et al. (2019) for NOEC chronic. However, it is considerably similar to the factors of 0.33, presented by De Zwart (2002) and Posthuma et al. (2019) respectively for NOEC chronic. For the transition from EC10eq acute to EC10eq chronic, this study factor is 0.3, closely mirroring the 0.32 from Aurisano et al. (2019) and 0.33 from De Zwart (2002) and Posthuma et al. (2019). Thus, the default generic extrapolation factors reported in the literature for EC50eq acute to EC10eq chronic are within a factor of 4.2 (0.25/0.06).
The differences observed between our results and those from previous studies can be attributed to several factors. One primary distinction lies in the aggregation process. While Aurisano et al. (2019) employed a presumably arithmetic average across datapoints for aggregation, Saouter, Wolff, et al. (2019) utilized the geometric mean. In our study, we adopted the geometric mean approach as detailed in Sect. 2.3. Additionally, the methodology for calculating extrapolation factors varied among studies. Aurisano et al. (2019) and Payet (2004) relied on regression analyses, considering both a free slope and a slope forced to unity, and incorporated a 95% confidence interval (CI) range. Conversely, Saouter, Wolff, et al. (2019) determined their factors using the geometric mean of the ratios of the compared effect concentration indicators. In our research, we employed regression analyses with a free slope for best-fit regression equations and a slope forced to unity for default factors. Another reason is the type of effect concentration indicators considered in different studies ranging from NOEC, EC10, and EC50, but in this study, we considered only EC10eq and EC50eq based on available knowledge and evidence for combining EC10 and NOEC to EC10eq.
Duplicate management would also have played a role. There is no mention of the removal of duplicates in Aurisano et al. (2019) whereas we eliminated approximately 8% of our initial dataset in the process of removing duplicates. While duplicate removal ought to increase the accuracy of the analysis, it will also have the effect of reducing the correlation coefficients associated with the extrapolation factors. There is a possibility that our approach may have eliminated some false positive matches, where apparently duplicate results reflect precise reproducibility in toxicological testing, but given the inherent variability of biological tests and our use of multiple duplicated metadata fields to identify duplicates, we consider our approach is likely to have eliminated more duplicate records than exactly reproduced toxicological results.
Perhaps, the most important factor is that, compared to earlier research, this study benefits from a significantly larger dataset. While Aurisano et al. (2019) had 9,627 datapoints, Payet (2004) featured 134,088, and Saouter, Wolff, et al. (2019) presented 54,353, our research encompasses 339,729 datapoints across 10,668 chemicals. Furthermore, in terms of effect concentration indicator and exposure duration combinations, our study uniquely offers two effect concentration indicators, EC10eq and EC50eq, paired with both acute and chronic exposure types. This approach provides the analyst with a wider range of options for extrapolation factors. Notably, our study delves into eight species groups, offering a more granular perspective on species group-specific extrapolation factors compared to prior studies that primarily focused on algae/cyanobacteria, crustaceans, and fish.
The primary aim of this study was to calculate extrapolation factors at various specific species and generic levels using a new harmonization of effect concentration indicators. Deriving extrapolation factors for individual chemical subgroups is a natural next step to enhance the accuracy of these factors. However, to avoid erroneous assignments, the precise definition of such subgroups based on chemical structure, known modes of action, or uses requires careful consideration. For example, in the PFAS grouping used in this study, a total of 202 chemicals were found in the freshwater ecosystem with available data. The R2 values were low due to the high diversity within the group, indicating that further classification into different PFAS subgroups may be necessary to increase coherence and improve R2 values.
Although this is beyond the scope of this study, we attempted to provide generic extrapolation factors for different chemical groups utilizing chemical groups based on the EPA DSSTox Tree. In total, generic extrapolation factors were calculated for 15 chemical groups as detailed in the supplementary information in Table G3. Due to limitations in the number of datapoints per group, species-specific extrapolation factors were not calculated, but generic factors were provided. The default extrapolation factors for different chemical groups to EC50eq chronic are within the range of 0.77 to 2.56 from EC10eq acute, 2.71 to 7.69 from EC10eq chronic, and 0.21 to 0.59 from EC50eq acute, with median values of 0.99, 3.48, and 0.42, respectively. These correspond to values of 0.99, 3.51, and 0.45 calculated for all organic chemicals in the freshwater ecosystem in this study. For extrapolation to EC10eq chronic, the factors range from 0.24 to 0.41 from EC10eq acute, 0.05 to 0.15 from EC50eq acute, and 0.13 to 0.37 from EC50eq chronic, with median values of 0.30, 0.11, and 0.29. These correspond to values of 0.29, 0.13, and 0.29 calculated for all organic chemicals in the freshwater ecosystem in this study. This indicates that future refinements in extrapolation factors for different chemical groups will not primarily result from the addition of new data, but also from a more detailed examination of already available data.
4 Conclusions
This paper presents three steps forward in relation to previous work: it uses a more extensive database of experimental ecotoxicity data than its predecessors accounted for 339,729 datapoints across 10,668 chemicals, it uses a modified curation process compared with them, and it delivers extrapolation factors converting various effect concentration indicators to EC10eq chronic that is recommended for calculating USEtox EFs. This research calculated 3 generic and 24 species group-specific extrapolation factors, tailored to various effect concentration indicators and exposure durations, facilitating the conversion of effect concentration indicator groups (EC10eq and EC50eq) to a chronic EC10eq. This, in turn, will enable ecotoxicity characterization factor calculations in USEtox. It is to be expected that more data will be available for calculations like ours in the future as toxicological testing and digitalization proceed. However, it is unlikely that large differences will occur as each individual new datapoint will have less and less impact on the end results. Thus, either looking specifically at now understudied groups or generating specifically more EC10eq chronic data should be prioritized.
Data availability
All data generated during this study are included in this published article and its supplementary information file. Data from REACH is available on the European Chemicals Agency (ECHA) website, while the CompTox Chemicals Dashboard data is accessible via the website of the United States Environmental Protection Agency (US EPA).
References
Adkins N (2023) COMPTOX_Public_ToxValDB v9.4 https://clowder.edap-cluster.com/datasets/61147fefe4b0856fdc65639b#folderId=645a5c0ce4b08a6b39438b10&page=0. Accessed 30 Jul 2023
Aggarwal R, Holmquist H, Arvidsson R, Reppas-Chrysovitsinos E, Peters G (2024) Influence of data selection on aquatic ecotoxicity characterization factors for selected persistent and mobile substances. Int J Life Cycle Assess 29(2):344–354. https://doi.org/10.1007/s11367-023-02263-w
Aurisano N, Albizzati PF, Hauschild M, Fantke P (2019) Extrapolation factors for characterizing freshwater ecotoxicity effects [article]. Environ Toxicol Chem 38(11):2568–2582. https://doi.org/10.1002/etc.4564
Aurisano N, Jolliet O, Chiu WA, Judson R, Jang S, Unnikrishnan A, Kosnik MB, Fantke P (2023) Probabilistic points of departure and reference doses for characterizing human noncancer and developmental/reproductive effects for 10,145 chemicals. Environmental Health Perspectives 131(3):037016. https://doi.org/10.1289/EHP11524
Azimonti, & et al (2015) Comparison of NOEC values to EC10/EC20 values, including confidence intervals, in aquatic and terrestrial ecotoxicological risk assessment. EFSA Supporting Publications, 12(12), 906E
Chang X, Tan Y-M, Allen DG, Bell S, Brown PC, Browning L, Ceger P, Gearhart J, Hakkinen PJ, Kabadi SV, Kleinstreuer NC, Lumen A, Matheson J, Paini A, Pangburn HA, Petersen EJ, Reinke EN, Ribeiro AJS, Sipes N, Mumtaz M (2022) IVIVE: facilitating the use of in vitro toxicity data in risk assessment and decision making. Toxics 10(5):232 (https://www.mdpi.com/2305-6304/10/5/232)
Crane M, Newman MC (2000) What level of effect is a no observed effect? Environ Toxicol Chem 19(2):516–519. https://doi.org/10.1002/etc.5620190234
De Zwart D (2002) Observed regularities in species sensitivity distributions for aquatic species. In: L Posthuma, GW Suter, TP Traas (eds) Species sensitivity distributions in ecotoxicology. CRC, Boca Raton, FL, USA, pp 157–178
Douziech M, Oginah SA, Golsteijn L, Hauschild MZ, Jolliet O, Owsianiak M, Posthuma L, Fantke P (2024) Characterizing freshwater ecotoxicity of more than 9000 chemicals by combining different levels of available measured test data with in silico predictions. Environ Toxicol Chem, n/a(n/a). https://doi.org/10.1002/etc.5929
ECHA (2008) Guidance on information requirements and chemical safety assessment. Chapter R.10: Characterisation of dose [concentration]-response for environment. (Risk characterisation, Issue. https://echa.europa.eu/guidance-documents/guidance-on-information-requirements-and-chemical-safety-assessment. Accessed 5 Dec 2023
ECHA (2012) Guidance on information requirements and chemical safety assessment Chapter R.8: Characterisation of dose [concentration]-response for human health (ECHA-2010-G-19-EN ). E. C. Agency. https://echa.europa.eu/documents/10162/17224/information_requirements_r8_en.pdf. Accessed 5 Dec 2023
ECHA. (2023). Pre-registered substances - ECHA. Retrieved March from https://echa.europa.eu/information-on-chemicals/pre-registered-substances. Accessed 5 Dec 2023
EFSA, P. o. P. P. P. t. R. (2013) Guidance on tiered risk assessment for plant protection products for aquatic organisms in edge‐of‐field surface waters. EFSA J, 11(7), 3290
US EPA (1991) Technical support document for water quality-based toxics control (EPA/505/2–90–001, Issue. Office of Ground Water and Drinking Water: Washington, DC, 1991. Accessed 15 August 2
EU, E. C. (2023). Environmental footprint methods. https://environment.ec.europa.eu/news/environmental-footprint-methods-2021-12-16_en. Accessed 19 Dec 2023
European Commission (2011) Technical guidance for deriving environmental quality standards. common implementation strategy for the water framework directive (2000/60/EC). Guidance document no. 27. Prepared by EU, Member States and stakeholders. Publications Office of the European Union. https://doi.org/10.2779/43816
Fantke P, Aurisano N, Bare J, Backhaus T, Bulle C, Chapman PM, De Zwart D, Dwyer R, Ernstoff A, Golsteijn L, Holmquist H, Jolliet O, McKone TE, Owsianiak M, Peijnenburg W, Posthuma L, Roos S, Saouter E, Schowanek D, Hauschild M (2018) Toward harmonizing ecotoxicity characterization in life cycle impact assessment. Environ Toxicol Chem 37(12):2955–2971. https://doi.org/10.1002/etc.4261
Fantke P, Aurisano N, Provoost J, Karamertzanis PG, Hauschild M (2020) Toward effective use of REACH data for science and policy. Environ Int 135:105336
Fantke P, Bijster M, Guignard C, Hauschild M, Huijbregts M, Jolliet O, Kounina A, Magaud V, Margni M, McKone TE, Posthuma L, Rosenbaum R, Van de meent D, Zelm R (2017) USEtox 2.0 : Documentation (Version 1). https://usetox.org/. Accessed 1 Sept 2023
Gustavsson MB, Hellohf A, Backhaus T (2017) Evaluating the environmental hazard of industrial chemicals from data collected during the REACH registration process. Sci Total Environ 586:658–665. https://doi.org/10.1016/j.scitotenv.2017.02.039
Hahn T, Diamond J, Dobson S, Howe P, Kielhorn J, Koennecker G, Lee-Steere C, Mangelsdorf I, Schneider U, Sugaya Y, Taylor K, Dam RV, Stauber JL (2014) Predicted no effect concentration derivation as a significant source of variability in environmental hazard assessments of chemicals in aquatic systems: an international analysis. Integr Environ Assess Manag 10(1):30–36. https://doi.org/10.1002/ieam.1473
Hauschild MZ, Huijbregts M, Jolliet O, Macleod M, Margni M, van de Meent D, Rosenbaum RK, McKone TE (2008) Building a model based on scientific consensus for life cycle impact assessment of chemicals: the search for harmony and parsimony. Environ Sci Technol 42(19):7032–7037. https://doi.org/10.1021/es703145t
ISO (2006) ISO14040:2006. Environmental management - Life cycle assessment - Principles and framework. International Organization for Standardization, ISO Central Secretariat. https://www.iso.org/standard/38498.html
Iwasaki Y, Kotani K, Kashiwada S, Masunaga S (2015) Does the choice of NOEC or EC10 affect the hazardous concentration for 5% of the species? Environ Sci Technol 49(15):9326–9330. https://doi.org/10.1021/acs.est.5b02069
Jacobs MM, Malloy TF, Tickner JA, Edwards S (2016) Alternatives assessment frameworks: research needs for the informed substitution of hazardous chemicals. Environ Health Perspect 124(3):265–280. https://doi.org/10.1289/ehp.1409581
Judson R (2018) ToxValDB: Compiling publicly available in vivo toxicity data. EPA's computational toxicology communities of practice monthly meeting, RTP, NC, December. https://doi.org/10.23645/epacomptox.7800653.v1
King OC, Smith RA, Mann RM, Warne MSJ (2017) Proposed aquatic ecosystem protection guideline values for pesticides commonly used in the Great Barrier Reef catchment area: Part 1 (amended) - 2,4-D, Ametryn, Diuron, Glyphosate, Hexazinone, Imazapic, Imidacloprid, Isoxaflutole, Metolachlor, Metribuzin, Metsulfuron-methyl, Simazine, Tebuthiuron. Department of Science, Information Technology and Innovation. Brisbane, Queensland, Australia. https://www.waterquality.gov.au/anz-guidelines/guidelinevalues/default/draft-dgvs#third-party-process-for-proposing-default-guideline-values. Accessed Dec 2023
Klimisch HJ, Andreae M, Tillmann U (1997) A systematic approach for evaluating the quality of experimental toxicological and ecotoxicological data. Regul Toxicol Pharmacol 25(1):1–5. https://doi.org/10.1006/rtph.1996.1076
Klok C, de Vries P, Jongbloed R, Tamis J (2012) Literature review on the sensitivity and exposure of marine and estuarine organisms to pesticides in comparison to corresponding fresh water species. EFSA Support Publ 9(11):357E. https://doi.org/10.2903/sp.efsa.2012.EN-357
Kristiansson E, Coria J, Gunnarsson L, Gustavsson M (2021) Does the scientific knowledge reflect the chemical diversity of environmental pollution? – a twenty-year perspective. Environ Sci Policy 126:90–98. https://doi.org/10.1016/j.envsci.2021.09.007
Länge R, Hutchinson TH, Scholz N, Solbé J (1998) Analysis of the ECETOC Aquatic Toxicity (EAT) database. II - Comparison of acute to chronic ratios for various aquatic organisms and chemical substances. Chemosphere 36(1):115–127. https://doi.org/10.1016/S0045-6535(97)10024-8
Lowe CN, Williams AJ (2021) Enabling high-throughput searches for multiple chemical data using the U.S.-EPA CompTox chemicals dashboard. J Chem Inf Model 61(2):565–570. https://doi.org/10.1021/acs.jcim.0c01273
Moore DRJ, Caux P-Y (1997) Estimating low toxic effects. Environ Toxicol Chem 16(4):794–801. https://doi.org/10.1002/etc.5620160425
Müller N, de Zwart D, Hauschild M, Kijko G, Fantke P (2017) Exploring REACH as a potential data source for characterizing ecotoxicity in life cycle assessment. Environ Toxicol Chem 36(2):492–500
Owsianiak M, Hauschild MZ, Posthuma L, Saouter E, Vijver MG, Backhaus T, Douziech M, Schlekat T, Fantke P (2023) Ecotoxicity characterization of chemicals: global recommendations and implementation in USEtox. Chemosphere 310:136807. https://doi.org/10.1016/j.chemosphere.2022.136807
Payet J (2004) Assessing toxic impacts on aquatic ecosystems in life cycle assessment (LCA). Ph.D. Dissertation, Institut des Sciences et Technologies de L'environment, Ecóle Polytechnique Fédérale de Lausanne, Lausanne, Switzerland. https://doi.org/10.1065/lca2005.09.003
Posthuma L, van Gils J, Zijp MC, van de Meent D, de Zwartd D (2019) Species sensitivity distributions for use in environmental protection, assessment, and management of aquatic ecosystems for 12 386 chemicals. Environ Toxicol Chem 38(4):703–711. https://doi.org/10.1002/etc.4373
REACH. (2020). REACH Dossier Database. Retrieved August from https://echa.europa.eu/fr/search-for-chemicals. Accessed Aug 2020
Rodea-Palomares I, Bone AJ (2024) Predictive value of the ToxCast/Tox21 high throughput toxicity screening data for approximating in vivo ecotoxicity endpoints and ecotoxicological risk in eco- surveillance applications. Sci Total Environ 914:169783. https://doi.org/10.1016/j.scitotenv.2023.169783
Rosenbaum RK, Bachmann TM, Gold LS, Huijbregts MAJ, Jolliet O, Juraske R, Koehler A, Larsen HF, MacLeod M, Margni M, McKone TE, Payet J, Schuhmacher M, van de Meent D, Hauschild MZ (2008) USEtox—the UNEP-SETAC toxicity model: recommended characterisation factors for human toxicity and freshwater ecotoxicity in life cycle impact assessment. Int J Life Cycle Assess 13(7):532–546. https://doi.org/10.1007/s11367-008-0038-4
Sala S, Biganzoli F, Mengual ES, Saouter E (2022) Toxicity impacts in the environmental footprint method: calculation principles. Int J Life Cycle Assess 27(4):587–602
Saouter E, Biganzoli F, Ceriani L, Versteeg D, Crenna E, Zampori L, Sala S, Pant R (2018) Environmental footprint: update of life cycle impact assessment methods-ecotoxicity freshwater, human toxicity cancer, and non-cancer. Publications Office of the European Union, Luxembourg. https://doi.org/10.2760/611799
Saouter E, Biganzoli F, Pant R, Sala S, Versteeg D (2019) Using REACH for the EU environmental footprint: building a usable ecotoxicity database Part I. Integr Environ Assess Manag 15(5):783–795. https://doi.org/10.1002/ieam.4168
Saouter E, Wolff D, Biganzoli F, Versteeg D (2019) Comparing options for deriving chemical ecotoxicity hazard values for the european union environmental footprint, Part II. Integr Environ Assess Manag 15(5):796–807. https://doi.org/10.1002/ieam.4169
Silva MH (2020) Use of computational toxicology (CompTox) tools to predict in vivo toxicity for risk assessment. Regul Toxicol Pharmacol 116:104724. https://doi.org/10.1016/j.yrtph.2020.104724
Tickner JA, Simon RV, Jacobs M, Pollard LD, van Bergen SK (2021) The nexus between alternatives assessment and green chemistry: supporting the development and adoption of safer chemicals. Green Chem Lett Rev 14(1):23–44. https://doi.org/10.1080/17518253.2020.1856427
U.S.EPA (2023) ECOTOX | Explore. Retrieved September from https://cfpub.epa.gov/ecotox/explore.cfm?sub=Species. Accessed 1 Sept 2023
USEPA (2023) ToxValDB v9.4 The United States Environmental Protection Agency’s Center for Computational Toxicology and Exposure. https://doi.org/10.23645/epacomptox.20394501.v5. Accessed 30 Jul 2023
Warne MS, Batley G, van Dam R, Chapman J, Fox D, Hickey C, Stauber J (2015) Revised method for deriving Australian and New Zealand water quality guideline values for toxicants. Prepared for the Council of Australian Government’s Standing Council on Environment and Water (SCEW), Department of Science, Information Technology and Innovation, Brisbane, QLD, Australia. https://www.waterquality.gov.au/anz-guidelines/guideline-values/derive/warne-method-derive. Accessed 30 Nov 2023
Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, Richard AM (2017) The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminformatics 9(1):61. https://doi.org/10.1186/s13321-017-0247-6
Williams AJ, Lambert JC, Thayer K, Dorne J-LCM (2021) Sourcing data on chemical properties and hazard data from the US-EPA CompTox Chemicals Dashboard: a practical guide for human risk assessment. Environ Int 154:106566. https://doi.org/10.1016/j.envint.2021.106566
Zhang Q, Li J, Middleton A, Bhattacharya S, Conolly RB (2018) Bridging the data gap from in vitro toxicity testing to chemical safety assessment through computational modeling [review]. Front Public Health, 6. https://doi.org/10.3389/fpubh.2018.00261
Funding
Open access funding provided by Chalmers University of Technology. This work is associated with the European Union Horizon 2020 research and innovation programme under grant agreement No 101036756, project ZeroPM: Zero Pollution of persistent, mobile substances.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Ralph K. Rosenbaum.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aggarwal, R., Gustavsson, M., Peters, G. et al. Extrapolation factors for calculating ecotoxicity effects in LCA. Int J Life Cycle Assess (2024). https://doi.org/10.1007/s11367-024-02357-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11367-024-02357-z