Abstract
Aging has become one of the fastest-growing research topics in biology. However, exactly how the aging process occurs remains unknown. Epigenetics plays a significant role, and several epigenetic interventions can modulate lifespan. This review will explore the interplay between epigenetics and aging, and how epigenetic reprogramming can be harnessed for age reversal. In vivo partial reprogramming holds great promise as a possible therapy, but several limitations remain. Rejuvenation by reprogramming is a young but rapidly expanding subfield in the biology of aging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For decades, aging has been the single best predictor of human mortality in developed countries [1]. It is the major risk factor for several of the top causes of death, such as cardiovascular disease and cancer [2,3,4]. Organismal aging also greatly enhances the susceptibility to chronic diseases, such as diabetes, neurodegeneration, and metabolic syndromes [5]. As a complex, multi-factorial biological process, it has been typically defined by the presence of specific hallmarks [6]. Those include loss of proteostasis, mitochondrial dysfunction, genomic instability, and epigenetic alterations. Nevertheless, multiple theories have been proposed to explain the mechanism behind the convoluted aging process, many relying on only one factor. The “cross-linkage theory of aging” explains the loss of proteostasis as being due to increased hazardous crosslinking between cellular proteins [7]. The “free radical theory of aging” posited that elevated reactive oxygen species (ROS) and the resultant accumulation of cellular damage are responsible for the aging phenotype [8, 9]. Somatic DNA damage and epigenetic modifications have also been at the core of other aging theories. The “information theory of aging,” proposed by David Sinclair in 2019, suggests that loss of epigenetic information through time, like a scratched vinyl disc, is the basis for age-associated cellular deterioration [10]. Even though no theory has been proved beyond doubt, mounting evidence indicates that specific modifications in epigenetic marks are responsible for cellular and organismal aging.
According to the NIH Epigenomics Roadmap Project, “epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable” [11]. Changes in histone variants, histone post-translational modifications (PTMs), DNA methylation (DNAm), among others affect gene expression and packing of chromatin, the DNA-protein complex, and fall under the field of epigenetics. These alterations are mediated by several enzymes that act as readers and modifiers, particularly histone methyltransferases (HMTs), demethylases (HDMTs), acetyltransferases (HATs), and deacetylases (HDACs). The cellular epigenetic state is a dynamic interplay of all these components and changes over time and with environmental stimuli.
Isogenic studies across species demonstrate the contributory role of epigenetics in aging. In budding yeast (Saccharomyces cerevisiae), early stochastic epigenetic changes markedly determine single-cell replicative lifespan [12]. Worker and queen bees possess the same genetic information, yet display strikingly different phenotypes and lifespan [13, 14]. In mice, a precocious aging phenotype can be induced by chemicals that disrupt epigenetic marks during development [15]. In humans, a similar divergence is observed with twin studies. Herskind et al. estimated that only about 25% of the variance in longevity could be attributed to the identical genetic makeup of monozygotic twins [16]. Epigenetic marks early in life are virtually identical, whereas they differ later on through so-called epigenetic drift [17]. Divergent epigenetic signatures can thus explain phenotypic differences between isogenic twins [18]. This epigenetic drift can be observed early in yeast. After an initial stochastic period, one of two epigenetic aging routes is committed to under the same genetic background and environment, resulting in a 50% replicative lifespan difference [12]. Overall, epigenetic variations are ubiquitous regulators of the aging process in various of organisms across several kingdoms.
The irreversibility of aging was assumed as recently as the end of the twentieth century [19], partly because DNA double-strand breaks and mutations, thought to be one of the causes of aging, accumulate with time [20,21,22,23,24]. However, genetic damage is not always correlated with aging [25]. Around that time, epigenetic modifiers such as yeast Sir2 were known to be implicated in aging, and their overexpression extended lifespan [26]. Most interventions, such as calorie restriction, slowed aging. With the discovery that aged, differentiated cells can be reversed to phenotypically young, embryonic-like stem cells [27], developmental reversal was shown to be attainable. This procedure is referred to as “epigenetic reprogramming.” Recent studies have begun to explore its use in inducing age reversal by modifying the epigenome [28, 29]. Nowadays, it is known that aging can be slowed, paused [30, 31], and even reversed.
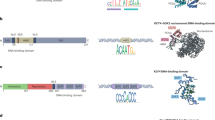
Recurrent imagery often used in developmental biology is the “epigenetic landscape” (Fig. 1 [32]), which facilitates the understanding of the aging process. After coining the term epigenetics as the “science concerned with the causal analysis of development,” Waddington created the landscape analogy in the middle of the twentieth century [32, 33]. It originally served to represent the initial stochasticity and later determinism of cell differentiation in organismal development with a marble rolling down a landscape with several valleys. The marble faces several branching points on the landscape, i.e., the choices for cell fate determination. The original epigenetic landscape is useful to visualize simple developmental pathways but limited since the topography is static. Hence, throughout this article, a modified landscape based on the malleable free-energy diagram will be used to explore the epigenetic modifications associated with aging and epigenetic reprogramming (Fig. 2). There are several local maxima and minima, with the height representing epigenetic instability and the location, a single epigenetic state. Going down the landscape can be imagined as losing epigenetic plasticity potential.
Drawing of the original epigenetic landscape proposed by Waddignton. Figure from [32]
Here, this article will briefly review the findings of age-related epigenetic changes from recent studies (for more information, see Kane et al. [34]). Then, epigenetic modifications driven by reprogramming will be discussed, alongside the potential to revert aging and its limitations. Finally, reprogramming will be explored alongside other well-established life-extension interventions.
Sliding down the epigenetic landscape: age-related epigenetic modifications
During mammalian development, the zygote is the first step in a series of preprogrammed events that result in a fully fledged adult. This single cell lies at the epigenetic landscape’s central and highest peak (Fig. 2). As time progresses, the zygote-marble descends the mountain through a succession of ever deeper valleys, ensuring cell fate stability. As the marble continues along its trajectory, several epigenetic modifications occur as it distances the center. Those include chromatin remodeling, differential histone PTMs, accumulation of histone variants, and regional hyper and hypo DNAm [3, 35].
Reduced global heterochromatin
A gradual, net loss of heterochromatin with advancing age, observed across organisms from yeast to mammals, has been the focus of the “heterochromatin loss model of aging” [36]. It proposes that a reduction in densely packed DNA is the culprit for age-related phenotypes. The chromatin’s elemental structural unit is the nucleosome, an octamer composed of histone proteins (canonically H2A, H2B, H3, and H4) with DNA wrapped around [37, 38]. It is the basis of a significant portion of epigenetic regulation.
Supranucleosomal chromatin organization plays a key role in gene expression [39]. Transcription is affected by distinct DNA accessibility due to specific local crowding conditions in the DNA microenvironment. Loss of heterochromatin has been associated with global transcription increase [40], but packing-density also controls the genomic information space, being positively correlated with intercellular transcriptional heterogeneity [39, 41]. Elevated chromatin scaling is characteristic of cancer cells [41]. These modelling studies highlight the importance of maintaining appropriate genome topology since several diseases, including aging, are connected to reduced heterochromatin [42, 43]. Increased differential transcription steadily appears in eukaryotes with aging, from yeast to humans [36, 44,45,46,47]. In fact, the anti-inflammatory drug Celecoxib, also an adjuvant that modifies chromatin scaling [39], has been shown to extend lifespan in C. elegans [48]. The study presents evidence that the extension was achieved through PDK-1 activity modulation, but another explanation is a differential expression of the protein based on macrogenomic chromatin regulation.
Several molecular events are responsible for less dense genome topology. Across the genome, H1 linker histones compact chromatin by binding to short DNA segments in between nucleosomes, effectively folding the DNA-protein complex [49]. Reversible phosphorylation of serine and threonine residues at the C-terminal tail of H1 histones are responsible for regulating H1 packing behavior [50]. Individuals with a deletion encompassing these residues in one of the multiple copies of the gene display phenotypic attributes of premature aging [50]. Overall, their fibroblasts show more nucleoid relaxation, less condensed chromosomes, and higher nucleolar instability than controls [50]. Loss of silencing of nucleolar ribosomal DNA (rDNA) is also known to promote aging in budding yeast [26, 51, 52]. It is partly caused by core histone protein reduction since roughly half is lost in yeast replicative aging [53, 54]. A similar decline has been detected in the worm Caenorhabditis elegans [55], fruit fly Drosophila melanogaster [46], human fibroblasts [56], and senescent human cells [57]. Overexpression of histone proteins increases chromatin compaction and organismal lifespan across species [54, 58]. Paradoxically, histone transcripts increase with aging due to heterochromatin reduction, but functional protein synthesis is further reduced, leading to a net loss of nucleosome occupancy [40]. The consequences range from DNA damage and chromosomal translocations to integration of hazardous nucleic acids into the nuclear genome, such as mitochondrial DNA (mtDNA) and transposons [40]. Nevertheless, more open chromatin is not always detrimental. A slight reduction of 15% in histones H2A and H2B expression decreases chromatin packing but extends replicative lifespan in yeast [59]. A cellular response mediates such alterations through TOR, an important nutrient-sensing pathway that regulates aging. Even though chromatin density is an important epigenetic factor, numerous other epigenetic factors are correspondingly influential during organismal aging.
Histone post-translational modifications
A wide range of chemical alterations occurs on histones, most notably acetylation and methylation [60]. Over 1000 different modifications can occur in the histone tails and histone globular domains [61]. This network of histone changes is highly complex and mediates the precise regulation of gene expression. Some factors that affect the distinct behavior of the modifications include the histone residue’s location, the histone’s gene position, and the composite contribution of multiple alterations.
In general, histone acetylation has been thought to facilitate gene expression and be more prevalent during aging because of elevated global transcription. Levels of histone H4 lysine 16 acetylation (H4K16ac) increase with aging in yeast and interventions that lower its abundance increase lifespan [53]. For instance, deletion of a component of the H4 HAT NuA4 promotes replicative longevity in yeast [62]. Deleting SAS2, another HAT that acetylates H4K16, extends yeast replicative lifespan [53]. Sir2, an HDAC known to promote longevity, deacetylates H4K16Ac and increases yeast lifespan [53]. As another example, H3K27ac is elevated in aged human skeletal muscle [63]. Targeting the translation of HAT p300 with short hairpin RNA extends replicative lifespan in human fibroblasts; overexpression of p300 shortens it [64]. However, hypoacetylation is not always beneficial. Loss of function of the H3 deacetylase complex Rpd3 delays aging in yeast [65] and in the fruit fly [66]. Moreover, global loss of H3K27ac is observed during aging in human and mouse brains, and in human hematopoietic cells [67, 68]. Age-upregulated genes lose H3K27ac at both promoters and gene bodies, whereas in age-downregulated genes only in the promoter, suggesting a suppressive effect of gene-body H3K27ac [67]. Sodium butyrate and suberanilohydroxamic acid, HDAC inhibitors that increase global H3K27ac, downregulated age-upregulated genes and upregulated age-downregulated genes, restoring homeostasis in the mouse brain. For some modifications, a precise level of histone acetylation is necessary for optimal longevity. For instance, H3K56Ac levels reduce with aging in yeast but increasing or decreasing H3K56Ac by deleting HATs (Hst3, Hst4) and HDACs (Rtt109) shortens lifespan and disrupts genomic stability in yeast [53, 54, 69]. It is worth highlighting how some of the age-related histone changes are species- and cell-type-dependent.
Histone methylation can cause both gene activation and silencing, with the former typically increasing and the latter decreasing with age [44]. Reduction of global H3K4 trimethylation (H3K4me3) marks, an indicator of transcriptional activation, through knockdown of HMTs prolongs lifespan, whereas knockdown of H3K4me3 HDMTs accelerates aging in C. elegans [70]. In old flies, H3K4me3 levels are altered [46]. On the other hand, silencing histone methylation marks, such as H3K36me3, H3K27me3, and H3K9me3, generally leads to age-related decline in organisms. H3K36me3 levels lower due to replicative aging in yeast [71] and H3K27me3 is depleted with aging in C. elegans and human cells [72]. An abrupt surge in the levels of H3K27me3 demethylase UTX-1 is highly associated with mortality in C. elegans [72]. Decreased H3K9me3 is observed in old flies compared to young flies [46], and a similar reduction occurs in mouse and human bone marrow stromal cells [73]. However, in fly heads specifically, the opposite is true [74]. In fact, a progeroid mouse model displayed increased levels of H3K9me3 and defective DNA repair within dense chromatin, and lowering H3K9me3 levels partly reversed the precocious aging phenotype [75]. The above studies highlight how different cell types display distinct age-related epigenetic modifications. Similarly to Cheng et al. [67], perhaps the location of the histone PTM within a gene might determine activating or repressive behavior that could explain the tissue variations. Further studies are necessary to elucidate the role of histone methylation marks in various positions across the genome.
Several other poorly understood histone marks display altered levels with aging. Formylation is the second most abundant histone lysine acylation in mice livers, only behind acetylation [76]. It more than doubles with aging [76]. Aliphatic acylations and advanced glycation end products (AGEs) in general increase by approximately 50%. AGEs disrupt chromatin organization, but the role of each modification in aging has not been well-documented [77]. Permanent oxidative stress markers skyrocket, particularly with the oxidation of methionine sulfoxide to methionine sulfone increasing by 5–10 fold [76]. Citrullination, believed to be involved in DNA repair, increases by approximately 50% [76]. Further research is required to understand the composite role of all these different histone modifications in gene regulation. Accumulated DNA damage might drive some of these changes [78], but it is unknown how histone PTM epigenetic information is lost.
Chromatin modifier changes
Highly sophisticated interactions among histone proteins, nucleosome remodeling complexes (NRCs), histone modifiers (methylases, acetylases, etc.), and transcription factors alter during aging. Variants of the canonical histone proteins regulate chromatin dynamics, from assisting packing to DNA repair [79, 80]. Histone variant H3.3, initially thought to have no functional significance, accumulates in mouse brains over time [81, 82] and appears to drive cellular senescence [83]. MacroH2A, a variant associated with both transcriptional activation [84] and repression [85], increases in mice, primates, and human fibroblasts with aging [86]. Nucleosome remodeling complexes also play a role. Nucleosome remodeling deacetylase (NuRD) malfunctions in aging [87]. Isw2 and Chd1, ATP-dependent NRCs, are detrimental to yeast replicative lifespan [62]. RNA interference of the NRC SWI/SNF abolishes longevity extension in some cases in C. elegans [88]. Chromatin modifiers, such as HATs, HDACs, HMTs, and HDMTs, also modulate aging [53, 54, 62, 64,65,66, 72]. Another component is a change in transcription factors, which play key roles in DNA accessibility and modification [89, 90]. For example, loss of the transcription factor Slug in mice causes an aged phenotype in vivo [91].
DNA methylation
Several dynamic modifications are present in the DNA of most eukaryotes and are relevant to aging. Among them, 5’-cytosine methylation (5mC) is the most frequent, typically occurring at locations of a cytosine followed by a guanine (CpG sites) [92]. Other unfamiliar cytosine additions also exist, such as hydroxymethylation (hmC), formylation (fC), carboxylation (caC), and 4’ methylation [93, 94]. Even 6’-adenine methylation has been observed [92, 94, 95]. 5mC has been assumed to dampen gene expression through steric hindrance of transcription factors, but it might be involved in nuanced raised expression depending on the position in the genome [96,97,98,99,100,101]. For the most part, 5mC methylation dwindles during mammalian aging [102,103,104,105,106,107,108], although some studies using modern techniques do not corroborate such global findings [109, 110]. Some specific, apparently important CpG sites are, in contrast, hypermethylated [111,112,113,114]. During development and aging, there is a methylation peak, since embryonic stem cells and old cells are hypomethylated. Accordingly, a surge in CpG 5mC has been shown in infants from 6 to 52 weeks of age [100]. Most importantly, age-related 5mC hyper- and hypomethylation is localized at particular genomic loci [111, 115, 116].
This differential hyper- and hypomethylation across the genome can be used to accurately predict age and mortality [101, 111, 117,118,119]. Machine learning methods allied with CpG epigenetic data were harnessed to create the so-called epigenetic clocks (see Horvath et al [120]). 353 CpG sites in Horvath’s clock and 71 in Hannum’s clock precisely calculate a person’s age with a median error of less than four years [101, 111]. Horvath’s clock is so accurate that embryonic stem cells, which are only present before birth, possess slightly negative age [111, 121]. Even the epigenetic drift can be quantified given the higher variance of DNAm age later in life [101]. DNA methylation is intrinsically related to aging, and its genome pattern is universal across eukaryotes [94]. Even then, age-related differential DNA methylation is not solely responsible for the aging phenotype. Some species such as the fruit fly virtually lack 5mC [122].
Non-coding RNAs
Non-coding RNAs (ncRNAs), long believed to arise from transcriptional errors, are key players in epigenetic regulation [123]. They finely modulate messenger RNA (mRNA) transcription, splicing, and degradation [124] and assist in the maintenance of proper genome topology [125]. They are ubiquitous in the human transcriptome. More than half of the human genome is transcribed [126, 127], giving rise to a multitude of transcripts. ncRNAs are both positively and negatively correlated with aging. Micro RNAs (miRNAs), a class of small ncRNAs, are generally downregulated in old compared to young eukaryotes [128,129,130], but some delay or accelerate the aging phenotype across species [52, 131,132,133,134]. Long ncRNAs can also be detrimental or beneficial for the aging phenotype. High Gas5 expression, a type of long ncRNA, is related to impaired learning in mice [135]. At the same time, overexpression of Sarrah, another ncRNA, improves cardiac function in mice [136]. Overall, increased transcription of most long ncRNAs is damaging because of elevated R-loop formation, a three-stranded nucleic acid structure [69, 137]. It is more prone to DNA damage and leads to cellular senescence. Some long ncRNAs can even form a DNA-DNA-RNA triple helix [136]. The role of ncRNAs in aging is becoming clearer with recent studies, but their role in epigenetic modulation is certainly crucial for the aging process.
Transposition
The “transposon theory of aging” posits that transposable elements (TEs), dubbed “jumping genes” for their excision and reintegration potentials, cause cellular degeneration and aging [138]. They are usually silenced during youth, but as heterochromatin is lost, they become activated. Chromatin packing deregulation directly elicits expression of transposable elements with age in fruit flies [58] and mice [139]. The transcript levels increase as the cellular mechanisms that suppress integration become insufficient to prevent it. Overexpression of genes that stabilize heterochromatin and lamivudine (a drug that targets the TE machinery component reverse transcriptase) restrains TEs and extends lifespan [58]. As expected, specific TEs are both silenced and expressed differentially [140]. Almost all have biased de novo insertions in the genome, but long interspaced repeat element 1 (LINE1), the most abundant human TE [141], is not generally affected by the presence of heterochromatin [142]. LINE1 activation contributes to age-related inflammation and cellular senescence [139]. Stavudine, a LINE1 reverse-transcriptase inhibitor, rescues the young inflammation profile in mice [143] and lamivudine partly inhibits the cellular senescence phenotype [139]. The stavudine-treated group had approximately a 30% lower DNAm age [143]. These results suggest that transposition is not merely correlated with age but one of the causes of aging.
Climbing back up the epigenetic landscape: reversing epigenetic modifications by reprogramming
The first notable experiment that showed that a differentiated somatic cell still contains all the necessary genetic information to produce an entirely new organism was somatic cell nuclear transfer (SCNT) [144]. The process of completely modifying a cell phenotype was named reprogramming. More recently, in 2006, it was shown that exogenous expression of the four factors Oct3/4, Sox2, Klf4, and c-Myc (OSKM) is sufficient to transform fibroblasts into induced pluripotent stem cells (iPSCs) [27]. Forced expression of the pluripotency factors modifies the landscape by effectively flipping the topography (Fig. 3). The central peak where the marble initially dwelled becomes an abyss. It pulls the marble towards the center again to reduce epigenetic instability, representing the loss of somatic identity and gain of pluripotency.
In order for reprogramming to work correctly, efficient activation of the pluripotency network is the only requirement. Although the original OSKM combination is still used today, the complete cocktail of factors is not essential. Earlier, removing any of the factors did not elicit reprogramming [27]. With advances in the culture medium, three out of the four was enough, albeit with a lower efficiency [145]. SK alone could reprogram highly proliferative differentiated cells [145]. With the use of the clustered regularly interspaced short palindromic repeats (CRISPR) system, sole Oct4 or Sox2 overexpression lead to activation of the pluripotency circuitry [146]. There is an initial stochastic phase followed by a hierarchical, deterministic activation of certain genes, with Sox2 appearing to be a central node [147]. Any combination of factors that induces the network and produces the correct levels of pluripotency proteins can successfully reprogram [27, 148, 149].
As will be explored below, several epigenetic marks are effectively reversed during reprogramming. This remarkable process is being studied as a potential therapy against aging. Nevertheless, it has several limitations. Low efficiency, formation of teratomas (an extremely aggressive form of cancer), and persistence of certain epigenetic marks are some of the barriers to be overcome.
Epigenetic changes during reprogramming
Given the high degree of similarity between embryonic stem cells (ESCs) and iPSCs [27, 150,151,152], it is expected that some changes that occur during reprogramming might be a simple reversal of age-related epigenetic modifications. A few alterations, such as global hypomethylation and heterochromatin loss, get exacerbated during reprogramming, whereas others, such as telomere attrition, are rewound.
A recent longitudinal analysis of the transcriptome of the reprogramming intermediates of mouse embryonic fibroblasts (MEFs) identified key steps in the process [153]. Principal-component analysis highlighted five stages. 5mC DNA methylation declined markedly in the first step and then steadily dropped by half until the final stage when, intriguingly, it increased, but not enough to reach pre-reprogramming levels. These results suggest a first wave of demethylation to erase the differentiated cell program that primarily targets promoters, enhancers, and upstream regulatory elements. It is accompanied by a second, more gradual demethylation wave, which mainly affects gene bodies. Finally, a rapid surge in methylation, which may surpass progenitor-level global methylation [154], most likely determines the pluripotency program in the last step. The most important demethylation targets are pluripotency genes and their respective regulatory regions [27, 155]. Some types of transposons are demethylated, such as LINEs, others remain methylated, such as intracisternal A particle elements, and active retroviruses are silenced [145, 153, 156]. All these methylation changes contribute to the slightly negative age of iPSCs in Horvath’s epigenetic clock [111, 121].
When it comes to 5hmC, there is a global increase when comparing iPSCs to human fibroblasts [157]. TET1, the main enzyme responsible for regulating hydroxymethylation, is activated early during reprogramming [153] and is present in higher levels in iPSCs [157]. It is required for proper erasure of the previous cellular program during reprogramming [153]. Roughly 85% of the differentially hydroxymethylated regions are hyper hydroxymethylated, mostly concentrated near telomeres [157]. Gene expression is inversely correlated to 5hmC in transcriptional start sites [157]. A consequence is both hypomethylation and hypo hydroxymethylation in the regulatory elements of pluripotency factors [157]. Nevertheless, 5mhC is not a mere consequence of the oxidation of previously 5mC DNA, as increased hydroxymethylation is found in regions of both hyper and hypomethylation [157]. These observations suggest that 5hmC plays a role in regulating the pluripotency circuitry.
Another age-related epigenetic change that becomes exacerbated in reprogramming is the loss of heterochromatin. In OSKM reprogramming, OSK are the pioneer factors to induce global opening of chromatin [147]. Such chromatin unpacking has been observed even with in vivo reprogramming in mice, as DAPI, a DNA staining compound, elicits a weaker, more delocalized signal [158]. Heterochromatin protein 1β (HP1β), involved in gene expression regulation and DNA repair, is much more mobile in iPSCs than human fibroblasts [159]. HP1β mobility can even be used as a proxy to track reprogramming progress [159]. Recent advances in 5C and high-throughput sequencing have allowed a finer look at genome architecture modifications [151]. Topologically associating domains (TADs) show striking differences before and after reprogramming [151, 160]. Sliding back down to the center of the epigenetic landscape involves breaking and reforming cell-type-specific patterns of 3D interactions, albeit not all of them [151]. The more iPSCs are passaged, the more they acquire ESCs-specific higher-order chromatin connectivity [160]. However, a finer resolution in TAD mapping shows that iPSCs subdomain connectivity is not completely ESC-like [151], even though bigger domains become virtually indistinguishable.
There are also several changes observed with histone proteins and histone PTMs. In the Oct4 and Nanog promoters, H3K9 methylation remained constant, but H3 acetylation was much higher, indicating activation of pluripotency factors [27]. H3K9me3 and H4K20me3, silencing marks, decline globally but particularly at pericentric repeats and telomeres [156, 158]. H3K4me3 and H3K27me3, activating and repressive marks, respectively, change in two waves [155]. Initially, H3K4me3 relocates to other regions and H3K27me3 increases globally, particularly in differentiated cell-type-specific genes. Later, H3K4me3 levels surge and H3K27me3 moves to other areas [155]. These changes are concomitant to the first 5mC demethylation wave and the second 5mC methylation increase, indicating the activation and suppression of different genes across time in reprogramming [153]. In addition to histone PTMs, all the canonical histones are upregulated [161]. Even histone variants are likely to be repositioned through nucleosome remodeling, as H3.3 shifts downstream from promoters [162, 163].
Reprogramming does not always completely erase the previous cellular program, with some epigenetic remnants maintained after the process. The first induction of iPSCs already demonstrated that epigenetic marks, such as 5mC DNA methylation and H3K9me3, are not fully reversed on the promoters of pluripotency genes to levels seen in ESCs [27]. Moreover, genome-wide hydroxymethylation analysis of iPSCs derived from human fibroblasts highlights the presence of 20 large-scale regions with enduring 5hmC [157]. Tissue-specific residual 5mC methylation remains in iPSCs and these methylation foci might interfere with differentiation into some cell types [154, 164]. Moreover, some age-related methylation marks remain in iPSCs derived from aged donors. Based on Horvath’s epigenetic clock, there is a correlation, although weak, of the progenitor cells’ age and the epigenetic age of iPSCs [121]. Reprogramming of old cells result in a 5% global methylation increase [121]. These studies suggest iPSCs are primed to return to their original state more easily, as the trail traced on the epigenetic landscape is always a two-way pathway. Nevertheless, this epigenetic memory in reprogramming is generally assumed to be insignificant [165].
Overall, reprogramming takes a different route from a simple sequential reversal of age-related epigenetic changes. For instance, DNA methylation and global heterochromatin, which peak during youth, do not pass through the same peak. Another detour is evidenced by fibroblasts reprogrammed with OSKM taking a longer route if Oct4 is present, potentially reacquiring fibroblast features [145]. Oct4, in this case, leads to a loss of imprinting, misregulation of polycomb targets, and epigenetic anomalies, meandering through valleys on the epigenetic landscape. Different pathways to the center of the landscape can be taken with distinct epigenetic changes, as local minima become local maxima and vice versa. For more information on epigenetic changes during aging, see review by Papp and Plath [166].
Partial epigenetic reprogramming
In order for epigenetic reprogramming to be harnessed for the treatment of aging, dedifferentiation must not occur. The marble cannot return to the center, as complete in vivo reprogramming is a dangerous process that might lead to severe health problems or death. Hence, partial reprogramming, i.e., reversing age-related epigenetic changes without pluripotency acquisition, is the only feasible use of this technique. It has already been shown that reprogramming cells to increase “stemness” without reaching pluripotency drastically reduces the aged phenotype [167]. There is a steady, dramatic decrease in DNAm age during reprogramming [168]. The literature on this topic is current but scant and is summarized in Table 1.
Sarkar et al. published an analysis of transient expression of reprogramming factors in aged human fibroblasts and endothelial cells [29]. They transfected OSKM plus Lin28 and Nanog (OSKMLN) mRNAs for four days and performed several assays two days later. Transcriptome analysis showed a clear similarity between treated and young cells without activation of the pluripotency network. Similar or higher levels of heterochromatic H3K9me3 were found, which is intriguing given that full reprogramming depletes that histone PTM [156, 158]. This same epigenetic change was seen in partial reprogramming in mice fibroblasts [28]. Protein levels of Sirt1, an enzyme that promotes longevity [169], and HP1γ, an isoform of HP1β, increased. β-galactosidase, a hallmark of senescence [170], and proinflammatory senescence-associated secretory phenotype decreased, as seen in [28] as well. Strikingly, even Horvath’s epigenetic clock calculated an age reduction on average of 3.4 years. These youthful restorations mostly endured for the next 4 and 6 days after the interruption, although more moderately. In mice fibroblasts, the prior phenotype slowly restored after 4 and 8 days of interruption [28].
Reik at al. recently conducted a similar experiment using human fibroblasts from patients between 38 and 53 years [171]. The researchers used lentiviral transduction to transport an OSKM cassette inducible by the antibiotic doxycycline and started the reprogramming process. After 10 to 17 days, doxycycline was removed and the morphological changes observed upon reprogramming were reversed. In the successfully partially reprogrammed cells, a median drop of about 30 years in both the transcriptional clock and Horvath’s DNAm clock was observed, a much sharper decrease than observed by Sarkar et al. [29]. The youthful phenotype, characterized as a restoration in H3K9me3 levels and increase in collagen production, remained for an unspecified amount of time. Notwithstanding such encouraging results, the fibroblasts did momentarily lose their morphology during the transient reprogramming.
Sarkar and Reik’s reports indicate great promise in partial reprogramming, but other studies reported more cautious results. It is not surprising that transient reprogramming results in transient rejuvenation. OSKML reprogramming in senescent human fibroblasts for 9 days—it takes 40 to reach pluripotency in this case—did decrease β-galactosidase and increased HP1β mobility [159]. However, at day 12, the previous senescence phenotype returned [159]. Another study analyzed reprogramming in human mesenchymal stromal cells maintained in the same cell-specific medium [172]. Compared to control cells, they entered replicative senescence simultaneously and showed the same levels of β-galactosidase and p16, another senescence-associated marker [173]. Transient expression might lead to cell-fate anomalies. Partial OSKM reprogramming in mature lung epithelial cells over three weeks generated a non-natural progenitor [174]. Differently from the mature lung epithelial cells, the generated cells were easily expanded but did not display pluripotency markers [174]. The novel cell type’s appearance might be explained by the passage through a local maximum on the epigenetic landscape that represents a normally inaccessible epigenetic state; with the partial reprogramming, the local maximum becomes a local minimum. These contrasting results might have occurred due to the use of different reprogramming methods, cells, culture mediums, and lengths of expression.
To date, the most startling study showing the potential of partial reprogramming in tackling aging was conducted by Ocampo et al. [28]. The researchers used a progeroid mouse line that can express OSKM in vivo when given doxycycline, in contrast to in vitro conditions of all previously mentioned studies. Two days of induction followed by five days of abstention did not cause cancer or activation of pluripotency factors besides OSKM. The treatment led to a striking lifespan extension of approximately 50%. Several age-associated phenotypes were reversed, including restoration of normal H3K9me3 and H4K20me3 levels and reduced senescence-associated β-galactosidase. Partial in vivo reprogramming may also confer tissue regeneration capacity not even observed in young animals. It appears that transient reprogramming greatly supports tissue regeneration following injury [28, 175], as partially reprogrammed cells can replenish tissues more faithfully [29]. If the expression of pluripotency factors is not immediate after the damage, broad regeneration does not occur [175]. Since phenotypic plasticity is correlated with greater chromatin scaling [39, 41], partial reprogramming, by briefly opening chromatin [159], promotes an epigenetic environment that is fruitful for tissue regeneration. Tissue damage stimulates phenotypic plasticity and reprogramming as well [176]. In vivo reprogramming holds great promise for age reversal and tissue regeneration.
Limitations
Even with state-of-the-art techniques, several limitations hinder the applicability of reprogramming to confront aging. Besides the pharmacological difficulties of delivering a system capable of expressing the pluripotency factors, low efficacy, high heterogeneity, and severe side effects are pressing problems.
Reprogramming outcomes vary widely based on the reprogramming method, tissue source, progenitor cell age, and the cellular environment. An analysis of multiple reprogramming methods revealed drastic differences upon expression of the same set of pluripotency factors [177]. There was no methylation variation across methods, but retroviral-derived iPSCs were 13.5% aneuploid while mRNA-derived iPSCs were only 2.3%. Clustering of transcriptomes based on reprogramming techniques showed that, in general, similar procedures group together [152]. Tissue source may prime iPSCs to differentiate into their progenitor cells [154, 164]. The age of the progenitor cells also leads to heterogeneous reprogramming. There is evidence that donor age does not influence efficiency (typically 0.01–0.1%) even though it alters the number of successful cultures, leading to an overall lower yield [121, 164, 178]. Donor age also affects chromosomal abnormalities, DNA damage response, and apoptosis [179]. Culture medium affects variability and efficiency as well. Exchanging the culture medium between good and bad cultures narrows the difference in efficiency by 60% [178]. The ratio of inflammatory cytokines in the environment is partly responsible for the variability [176, 178], and might explain the increased heterogeneity in cultures of old progenitor cells. NF-κ B, a protein involved in cytokine production, and interferon-gamma, a cytokine, are likely reprogramming barriers [180]. Another reason for high heterogeneity in reprogramming is the differential epigenetic signature in pluripotency loci [181]. Precise placement of epigenetic modifiers where transcription is needed can greatly improve reprogramming [181, 182], as a valley is carved on the epigenetic landscape leading straight to the center (Fig. 4). Nevertheless, low efficiency and high heterogeneity can be solved with an improved cocktail of reprogramming factors and miRNAs. Efficiency of over 800% has been achieved since iPSC generation efficiency is calculated as the number of colonies generated per progenitor cells [183]. In single-cell experiments, 90.7% were successful. These results highlight the difficulty in generating reprogrammed cells, even though current protocols are close to converting 100% of the cells to iPSCs.
Full in vivo reprogramming is severely detrimental to health, leading to death or cancer development in a matter of days. Continuous expression of OSKM in mice results in death in a week caused by generalized abnormal tissue growth [28, 184, 185]. Reduced continuous expression still leads to death due to teratoma formation within weeks [184,185,186,187]. Nevertheless, the oncogenic potential of in vivo reprogramming does not likely arise from new mutations, as reprogramming does not appear significantly mutagenic [179, 188]. Shorter OSKM or OSK induction (fewer than five days) does not seem to result in cancer, even though temporary abnormal tissue growth and development is observed [185, 186]. These results suggest that a loss of cell-specific phenotype precedes cancer development. Transient OSKM or OSK expression also increases, for instance, colon cancer metastases [189]. A possible reason for the increased metastatic potential is the reshaping of the epigenetic landscape, with loss of tissue-specific transcription observed in multiple metastatic cancers [190]. Interestingly, several tumor suppressor genes function as barriers to reprogramming. p53, a protein in which mutations cause several types of cancer, decreases reprogramming efficiency [156, 176, 186, 191]. The tumor suppressor locus Ink4a/Arf codes for proteins that hinder reprogramming in vitro [192], even though it seems to be necessary for proper in vivo reprogramming [176]. Another complication of using reprogramming to tackle aging is that it triggers and is stimulated by cellular senescence, one of the hallmarks of aging [6]. In regions where in vivo reprogramming is successful, there is an increase in the number of β-galactosidase positive cells and the microenvironment is filled with cytokines and other senescence-associated molecules [176, 186]. Interestingly, reprogramming leads to both cancer cells, which have an epigenetic age much higher than the organism [111], and iPSCs, which are much younger, evidence indicating the flattening of the epigenetic landscape. All these results indicate that reprogramming indeed flattens and flips the epigenetic landscape, making it easier for the marble to take other pathways, including senescence and tumorigenesis. It is difficult—if not impossible—to control the boundaries precisely, as tissue type, age, and the cellular microenvironment affect reprogramming. It is worth noting, however, that a reduction in senescent cells was seen with cyclic partial reprogramming [28].
Another limitation in reprogramming to tackle aging is what can be described as the age-reversal-age-extension (Arae) paradox. In the epigenetic landscape, several lifespan-extending interventions that promote genomic stability make the valleys deeper. However, reprogramming flattens the landscape so that the marble can move back towards the center, making them intrinsically incompatible (Figs. 5 and 6). It has been shown that heterochromatin pathways act as barriers to reprogramming [193, 194]. The naked mole-rat (NMR) is an extremely long-lived murine, mainly because of its remarkable genomic stability. Reprogramming of NMR fibroblasts requires a special culture medium that quite easily reprograms mouse fibroblasts [195]. Three of the most well-known longevity pathways are AMPK, mTOR, and SIRT1. Metformin, thought to be an AMPK activator, increases lifespan across species [196,197,198,199]. In human cells, a therapeutic concentration of 100 μ M [200] was sufficient to alleviate several aging markers [201]. Only 10 μ M already significantly reduced reprogramming efficiency, and this inhibition increased with concentration [202]. Rapamycin, an inhibitor of mTOR, slows aging [203,204,205]. The drug appears to increase reprogramming potential at low doses with a peak at 0.3 nM [206], but decrease from 1 to 20 nM in a dose-dependent manner [207]. Human physiological concentrations range from 5 to 30 nM [208] and approximately 50 nM was used in studies showing lifespan extension [209, 210]. Resveratrol, involved in the SIRT1 pathway, ameliorates several age-associated phenotypes [211]. Even though it enhances reprogramming at 0.2–5 μ M, the effect wanes at 10 μ M and efficiency is drastically reduced at 20 μ M [212, 213]. Nevertheless, concentrations of even 500 μ M extended lifespan in yeast [214], and most of the health benefits of resveratrol are conferred in the 10–25 μ M range in vitro [215]. On the other hand, nicotinamide, a SIRT1 inhibitor, promotes reprogramming at the same concentration it reduces yeast replicative lifespan [191, 216]. The general enhancement of reprogramming efficiency at low doses of longevity-promoting compounds might be driven by increased epigenetic remodeling, since all the aforementioned drugs affect some epigenetic modifications. However, at higher doses that more optimally extend lifespan, the elevated genomic stability might hinder reprogramming. In summary, the essence of the Arae paradox is that several interventions that slow aging can be barriers to reprogramming.
Interventions that improve genomic stability also hinder reprogramming by enhancing the barriers to reach the center in comparison to Fig. 3
Concluding remarks
Epigenetics and aging are intrinsically related. Epigenetic modifications predictably occur throughout development and aging. Reduced global heterochromatin, altered histone PTMs, differential DNA methylation, and others account for the bulk of age-related changes. Targeting any one of these changes ameliorates aging hallmarks and extends lifespan. Epigenetic reprogramming reverses most if not all of the age-related epigenetic modifications. Harnessing partial reprogramming seems the most promising therapy to treat aging.
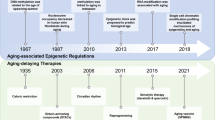
Even though the bulk of research concerning reprogramming only occurred in the last decade, several major strides have been made. Nowadays, we know how physical cues may assist in the process [217]. We can avoid heightened genome instability in iPSCs derived from aged progenitors [179]. In vivo delivery of pluripotency factors is vastly improving [187], as well as tissue regeneration by reprogramming [175, 183]. Even the power of CRISPR is being used in reprogramming [146, 149, 181, 182, 218].
There are still many limitations (Fig. 7). Whole organism delivery and homogeneous expression of pluripotency factors are challenging, currently unsolved tasks. Moreover, even though efficiency is greatly improving [183], most of the studies on reprogramming and aging still use only OSKM. Further research is also required to clarify the fine line between cancer and reprogramming. Lastly, the Arae paradox makes it unlikely that current lifespan interventions would have an additive effect alongside reprogramming.
A suggestion is an approach similar to the one shown by Ocampo et al. [28], but with the inclusion of a treatment that enhances genome stability during the absence of pluripotency factor expression. Short expression of pluripotency factors followed by administration of metformin, rapamycin, or even resveratrol would slightly flip the epigenetic landscape followed by the formation of deeper grooves. Perhaps this procedure would both improve lifespan extension and hinder the development of cancer and senescent cells. Further research is needed to check the validity of this speculative regimen.
Other combinations of reprogramming factors besides OSKM might prove more useful in decoupling age reversal from developmental reversal. It appears that the steady decline in DNAm age occurs before full reprogramming is achieved [168]. However, given the nonhomogeneous environment in vivo, there is a high risk that some cells will pass this threshold while others will likely not even experience epigenetic rejuvenation. Given the current knowledge of the pluripotency network [147], perhaps a cocktail of early reprogramming factors with late reprogramming inhibitors might reverse the aging phenotype without change of cell fate. Other well-known reprogramming barriers, such as p53, might be a barrier to full but not partial reprogramming. Further research ought to elucidate how to partially reprogram without cell dedifferentiation.
Other non-reprogramming strategies such as the use of drugs and young blood plasma are also proving promising for rejuvenation. A cocktail of metformin, human growth hormone, and dehydroepiandrosterone, rewound Horvath’s epigenetic clock by 2.5 years after one year of treatment in humans [219]. Moreover, it is known that parabiosis (the anatomical joining of two individuals) of old and young rats improves organ function in the aged animals [220]. It has recently been shown that administering young blood plasma to old rats more than halves the DNAm epigenetic age [221]. Such treatments can indirectly result in epigenetic modifications, yet are safer and less invasive than direct reprogramming. Non-reprogramming strategies might influence epigenetic marks by affecting the complex cellular regulatory networks downstream of gene expression. Because of their likely indirect effect, such treatments may not be as effective in age reversal as reprogramming; they might act as a stopgap before the challenging limitations of safe, effective in vivo reprogramming are resolved.
Aging reversal by epigenetic reprogramming is a new field of research, and not much has been studied. Nevertheless, even with all the current limitations, the future of reprogramming holds promise for the treatment of aging.
References
Harman D (1991) The aging process: major risk factor for disease and death. Proc Natl Acad Sci USA 88(12):5360–5363
Moskalev AA, Aliper AM, Smit-McBride Z, Buzdin A, Zhavoronkov A (2014) Genetics and epigenetics of aging and longevity, vol 13. Taylor and Francis , New York
Feser, J, Tyler, J. Chromatin structure as a mediator of aging. FEBS Lett, 2011; 585.
Brunet A, Berger SL (2014) Epigenetics of aging and aging-related disease. J Gerontol A Biol Sci Med Sci 69(S1):17–20
Kennedy B, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F (2014) Geroscience: linking aging to chronic disease, vol 159. Cell Press, Cambridge
López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging, vol 153. Cell Press, Cambridge
Bjorksten J (1968) The crosslinkage theory of aging, vol 16. Blackwell, Oxford
Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11(3):298–300
DENHAM HARMAN (1972) The biologic clock: the mitochondria? J Am Geriatr Soc 20(4):145–147
Sinclair D, LaPlante MD, Delphia C (2019) Lifespan : why we age– and why we don’t have to. ISBN 1-5011-9197-7
Roadmap Epigenomics Project - Overview
Jin M, Li Y, O’Laughlin R, Bittihn P, Pillus L, Tsimring LS, Hasty J, Hao N (2019) Divergent aging of isogenic yeast cells revealed through single-cell phenotypic dynamics. Cell Syst 8(3):242–253
Sargent M (2010) Why twins age differently. Nature 464(7292):1130–1131
Kucharski R, Maleszka J, Foret S, Maleszka R (2008) Nutritional control of reproductive status in honeybees via DNA methylation. Science 319(5871):1827–1830
Treviño LS, Dong J, Kaushal A, Katz TA, Jangid RK, Robertson MJ, Grimm SL, Ambati CSR, Putluri V, Cox AR, Kim K, May TD, Gallo MR, Moore DD, Hartig SM, Foulds CE, Putluri N, Coarfa C, LynWalker C (2020) Epigenome environment interactions accelerate epigenomic aging and unlock metabolically restricted epigenetic reprogramming in adulthood. Nat Commun 11(1):2316
Herskind AM, McGue M, Holm NV, Sørensen TIA, Harvald B, Vaupel JW (1996) The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet 97(3):319–323
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102(30):10604–10609
Poulsen P, Esteller M, Vaag A, Fraga MF The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007; 61(5 PART 2 SUPPL.)
Chow M, Rubin H (1996) Irreversibility of cellular aging and neoplastic transformation: a clonal analysis. Proc Natl Acad Sci USA 93(18):9793–9798
Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC (2004) Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol 6(2):168–170
Sedelnikova OA, Horikawa I, Redon C, Nakamura A, Zimonjic DB, Popescu NC, Bonner WM (2008) Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell 7(1):89–100
Liu B, Yip RKH, Zhou Z (2012) Chromatin remodeling, DNA damage repair and aging. Curr Genomics 13(7):533–547
Vijg J, Dollé MET (2002) Large genome rearrangements as a primary cause of aging. Mech Ageing Dev 123(8):907–915
Garcia AM, Calder RB, Dollé M. E. T., Lundell M, Kapahi P, Vijg J (2010) Age- and temperature-dependent somatic mutation accumulation in drosophila melanogaster. PLoS Genet 6(5):e1000950
Kaya A, Lobanov AV, Gladyshev VN (2015) Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae. Aging Cell 14(3):366–371
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles - a cause of aging in yeast. Cell 91(7):1033–1042
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Mo L i, Lam D, Kurita M, Beyret E, Araoka T, Vazquez-Ferrer E, Donoso D, Roman JL, Jinna X u, Esteban CR, Nuñez G., Delicado EN, Campistol JM, Guillen I, Guillen P, Belmonte JCI (2016) In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167(7):1719–1733
Sarkar TJ, Quarta M, Mukherjee S, Colville A, Paine P, Doan L, Tran CM, Chu CR, Horvath S, Qi L. e. i. S., Bhutani N, Rando TA, Sebastiano V (2020) Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat Commun 11(1):1–12
Hu CK, Wang W, Brind’Amour J, Singh PP, Adam Reeves G, Lorincz MC, Alvarado AS, Brunet A (2020) Vertebrate diapause preserves organisms long term through Polycomb complex members. Science 367(6480):870–874
Hussein AM, Wang Y, Mathieu J, Margaretha L, Song C, Jones DC, Cavanaugh C, Miklas JW, Mahen E, Showalter MR, Ruzzo WL, Fiehn O, Ware CB, Anthony Blau C, Ruohola-Baker H (2020) Metabolic control over mTOR-dependent diapause-like state. Dev Cell 52(2):236–250
Waddington CH (1957) The strategy of the genes; a discussion of some aspects of theoretical biology. Allen & Unwin, London
Baedke J (2013) The epigenetic landscape in the course of time: Conrad Hal Waddington’s methodological impact on the life sciences. Stud Hist Philos Biol Biomed Sci 44(4):756–773
Kane AE, David A (2019) Sinclair epigenetic changes during aging and their reprogramming potential, vol 54. Taylor and Francis Ltd, New York
O’Sullivan RJ, Karlseder J The great unravelling: chromatin as a modulator of the aging process. Trends Biochem Sci 2012;37
Bryant V (1997) The heterochromatin loss model of aging. In: Experimental gerontology, vol 32. Pergamon, pp 383–394
Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184(4139):868–871
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 n AA resolution. Nature 389(6648):251–260
Almassalha LM, Bauer GM, Wu W, Cherkezyan L, Di Z, Kendra A, Gladstein S, Chandler JE, Vanderway D, Seagle BLL, Ugolkov A, Billadeau DD, O’Halloran TV, Mazar AP, Roy HK, Szleifer I, Shahabi S, Backman V (2017) Macrogenomic engineering via modulation of the scaling of chromatin packing density. Nat Biomed Eng 1(11):902–913
Hu Zheng, Chen K, Xia Z, Chavez M, Pal S, Ja HS, Chen C, Li W, Tyler JK (2014) Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 28(4):396–408
RKA V, Wu W, Almassalha LM, Bauer GM, Li Y, VanDerway D, Frederick J, Di Z, Eshein A, Roy HK, Szleifer I, Backman V (2020) Disordered chromatin packing regulates phenotypic plasticity, vol 6
Zhang W, Li J, Suzuki K, Jing Q u, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Mo L i, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JCI (2015) A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348 (6239):1160–1163
Ren R, Deng L, Xue Y, Suzuki K, Zhang W, Yu Y, Wu J, Sun L, Gong X, Luan H, Yang F, Ju Z, Ren X, Si W, Tang H, Geng L, Zhang W, Li J, Qiao J, Xu T, Qu J, Liu GH (2017) Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res 27(4):483–504
McCauley BS, Dang W (2014) Histone methylation and aging: lessons learned from model systems, vol 1839. Elsevier, Amsterdam
Smeal T, Claus J, Kennedy B, Cole F, Guarente L (1996) Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84(4):633–642
Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li W Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8(1)
Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J (2005) Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci U.S.A 102(46):16690–16695
Ching TT, Chiang WC, Chen CS, Ao LH (2011) Celecoxib extends C. elegans lifespan via inhibition of insulin-like signaling but not cyclooxygenase-2 activity. Aging Cell 10(3):506–519
Hergeth SP, Schneider R (2015) The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 16(11):1439–1453
Flex E, Martinelli S, Dijck AV, Ciolfi A, Cecchetti S, Coluzzi E, Pannone L, Andreoli C, Radio FC, Pizzi S, Carpentieri G, Bruselles A, Catanzaro G, Pedace L, Miele E, Carcarino E, Ge X, Chijiwa C, Lewis MES, Meuwissen M, Kenis S, Van der Aa N, Larson A, Brown K, Wasserstein MP, Skotko BG, Begtrup A, Person R, Karayiorgou M, Louw Roos J, Van Gassen KL, Koopmans M, Bijlsma EK, Santen GWE, Barge-Schaapveld DQCM, Ruivenkamp CAL, Hoffer MJV, Lalani SR, Streff H, Craigen WJ, Graham BH, van den Elzen APM, Kamphuis DJ, Õunap K., Reinson K, Pajusalu S, Wojcik MH, Viberti C, Gaetano CD, Bertini E, Petrucci S, Luca AD, Rota R, Ferretti E, Matullo G, Dallapiccola B, Sgura A, Walkiewicz M, Frank Kooy R, Tartaglia M (2019) Aberrant function of the c-terminal tail of HIST1H1E accelerates cellular senescence and causes premature aging. Am J Hum Genet 105(3):493–508
Takehiko K Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci. 2011; 68
Saka K, Ide S, Ganley ARD, Kobayashi T (2013) Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr Biol 23(18):1794–1798
Dang W, Steffen KK, Perry R, Dorsey J, Johnson BF, Shilatifard A, Kaeberlein M, Kennedy B, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459(7248):802–807
Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK (2010) Elevated histone expression promotes life span extension. Mol Cell 39(5):724–735
Ni Z, Ebata A, Alipanahiramandi E, Lee SS (2012) Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 11(2):315–325
O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J (2010) Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17 (10):1218–1225
Adams PD, Ivanov A, Pawlikowski J, Manoharan I, Tuyn JV, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, Hong W u, Berger SL (2013) Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202(1):129–143
Wood JG, Jones BC, Jiang N, Chang C, Hosier S, Wickremesinghe P, Garcia M, Hartnett DA, Burhenn L, Neretti N, Helfand SL (2016) Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc Natl Acad Sci U.S.A 113(40):11277–11282
Yu R, Sun L, Yu S, Han X, Qin L, Dang W (2019) Cellular response to moderate chromatin architectural defects promotes longevity, vol 5
He H, Sabari BR, Garcia BA, Allis DC, Zhao Y (2014) SnapShot: histone modifications. Cell e1(2):458–458
Tony K (2007) Chromatin modifications and their function. Cell 128(4):693–705
McCormick MA, Delaney J. o. e. R., Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou ACZ, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, Bennett CF, Wang AM, Olsen B, Beyer RP, Bammler TK, Prunkard D, Johnson SC, Pennypacker JK, An E, Anies A, Castanza AS, Choi E, Dang N, Enerio S, Fletcher M, Fox L, Goswami S, Higgins SA, Holmberg MA, Hu D, Hui J, Jelic M, Ki SJ, Johnston E, Kerr EO, Kim J, Kim D, Kirkland K, Klum S, Kotireddy S, Liao E, Lim M, Lin MS, Lo WC, Lockshon D, Miller HA, Moller RM, Muller B, Oakes J, Pak DN, Peng ZJ, Pham KM, Pollard TG, Pradeep P, Pruett D, Rai D, Robison B, Rodriguez AA, Ros B, Sage M, Singh MK, Smith ED, Snead K, Solanky A, Spector BL, Steffen KK, Tchao BN, Ting MK, Wende HV, Wang D, Linnea Welton K, Westman EA, Brem RB, Liu XG, Suh Y, Zhou Z, Kaeberlein M, Kennedy B (2015) A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab 22(5):895–906
Zhou J, So KK, Li Y, Li Y, Yuan J, Ding Y, Chen F, Huang Y, Liu J, Lee W, Li G, Ju Z, Sun H, Wang H Elevated H3K27ac in aged skeletal muscle leads to increase in extracellular matrix and fibrogenic conversion of muscle satellite cells. Aging Cell. 2019;18(5)
Sen P, Lan Y, Li CY, Sidoli S, Donahue G, Dou Z, Frederick B, Chen Q, Luense LJ, Garcia BA, Dang W, Johnson BF, Adams PD, Schultz DC, Berger SL (2019) Histone acetyltransferase p300 induces de novo super-enhancers to drive cellular senescence. Mol Cell 73(4):684–698
Kim S, Benguria A, Lai CY, Jazwinski MS (1999) Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell 10(10):3125–3136
Rogina B, Helfand SL, Frankel S (2002) Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 298(5599):1745
Cheng H, Xuan H, Green CD, Han Y, Na S, Shen H, McDermott J, Bennett DA, Lan F, Han JDJ (2018) Repression of human and mouse brain inflammaging transcriptome by broad gene-body histone hyperacetylation. Proc Natl Acad Sci U.S.A 115(29):7611–7616
Adelman ER, Huang H-T, Roisman A, Olsson A, Colaprico A, Qin T, Lindsley CR, Bejar R, Salomonis N, Grimes LH, Figueroa ME (2019) Aging human hematopoietic stem cells manifest profound epigenetic reprogramming of enhancers that may predispose to leukemia. Cancer Discov 9(8):1080–1101
Feldman JL, Peterson CL (2019) Yeast sirtuin family members maintain transcription homeostasis to ensure genome stability. Cell Rep 27(10):2978–2989
Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS A systematic RNAi screen for longevity genes in C Elegans. Genes Dev. 2005;19
Sen P, Dang W, Donahue G, Dai J, Dorsey J, Cao X, Liu W, Cao K, Perry R, Lee J, Wasko BM, Carr DT, He C, Robison B, Wagner J, Gregory BD, Kaeberlein M, Kennedy B, Boeke JD, Berger SL (2015) H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev 29(13):1362–1376
Jin C, Li J, Green CD, Yu X, Tang X, Han D, Bo X, Wang D, Huang X, Cao X, Yan Z, Hou L, Liu J, Shukeir N, Khaitovich P, Chen CD, Zhang H, Jenuwein T, Han JDJ (2011) Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab 14(2):161–172
Huang B, Wang B, Lee WY-W, U KP, Leung KT, Li X, Liu Z, Chen R, Lin JC, Tsang LL, Liu B, Ruan Y, Chan HC, Gang Li, Jiang X (2019) KDM3A and KDM4C regulate mesenchymal stromal cell senescence and bone aging via condensin-mediated heterochromatin reorganization. iScience 21:375–390
Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, Neretti N, Helfand SL (2010) Chromatin remodeling in the aging genome of Drosophila. Aging Cell 9(6):971–978
Liu B, Wang Z, Le Z, Ghosh S, Zheng H, Zhou Z Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat Commun. 2013;4
Baldensperger T, Eggen M, Kappen J, Winterhalter PR, Pfirrmann T, Glomb MA (2020) Comprehensive analysis of posttranslational protein modifications in aging of subcellular compartments. Sci Rep 10(1):1–11
Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Bo L, Chandarlapaty S, Liu S, David Y Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun. 2019;10(1)
Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, Iacovoni JS, Rowicka M, Ginalski K, Côté J, Legube G (2018) Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol Cell 72(2):250–262
Skene PJ, Henikoff S Histone variants in pluripotency and disease. Development. 2013;140
Rohinton T Kamakaka and sue biggins Histone variants: deviants? Genes Dev. 2005;19
Piña B, Suau P (1987) Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev Biol 123(1):51–58
Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, Elsässer SJ, Guo Y, Ionete C, Hurd YL, Tamminga CA, Halene T, Farrelly L, Soshnev AA, Wen D, Rafii S, Birtwistle MR, Akbarian S, Buchholz BA, Blitzer RD, Nestler EJ, Yuan ZF, Garcia BA, Li S, Molina H, David Allis C (2015) Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87(1):77–94
Duarte LF, Young ARJ, Wang Z, Wu HA, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma’ayan A, Narita M, Bernstein E Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun. 2014;5
Chen H, Ruiz PD, McKimpson WM, Novikov L, Kitsis RN, Gamble MJ (2015) MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol Cell 59(5):719–731
Pasque V, Halley-Stott RP, Gillich A, Garrett N, Gurdon JB (2011) Epigenetic stability of repressed states involving the histone variant macroH2A revealed by nuclear transfer to Xenopus oocytes. Nucleus 2(6):533–539
Kreiling JA, Tamamori-Adachi M, Sexton AN, Jeyapalan JC, Munoz-Najar U, Peterson AL, Manivannan J, Rogers ES, Pchelintsev NA, Adams PD, Sedivy JM (2011) Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell 10(2):292–304
Pegoraro G, Kubben N, Wickert U, Göhler H, Hoffmann K, Misteli T (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol 11 (10):1261–1267
Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, Ruvkun G (2013) DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 15(5):491–501
Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, Grimm JB, Morisaki T (2016) Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165(3):593–605
Schübeler D (2015) Function and information content of DNA Methylation, vol 517. Nature Publishing Group, New York
Gross KM, Zhou W, Breindel JL, Ouyang J, Jin DX, Sokol ES, Gupta PB, Huber K, Zou L, Kuperwasser C (2019) Loss of slug compromises DNA damage repair and accelerates stem cell aging in mammary epithelium. Cell Rep 28(2):394–407
Lister R, Pelizzola M, Dowen RH, David Hawkins R, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Harvey Millar A, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462(7271):315–322
Aristizabal MJ, Anreiter I, Halldorsdottir T, Odgers CL, McDade TW, Goldenberg A, Mostafavi S, Kobor MS, Binder EB, Sokolowski MB, O’Donnell KJ Biological embedding of experience: a primer on epigenetics. Proc Natl Acad Sci. 2019; 201820838
Aliaga B, Bulla I, Mouahid G, Duval D, Grunau C (2019) Universality of the DNA methylation codes in Eucaryotes. Sci Rep 9(1):1–11
Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu J, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Margarita Behrens M, Ecker JR (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341:6146
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10):1010–1022
Lea AJ, Vockley CM, Johnston RA, Del Carpio CA, Barreiro LB, Timothy ER, Tung J Genome-wide quantification of the effects of DNA methylation on human gene regulation. eLife. 2018;7
Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, Nitta KR, Taipale M, Popov A, Ginno PA, Domcke S, Yan J, Schübeler D, Vinson C, Taipale J (2017) Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356:6337
Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS (2012) Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U.S.A 109(SUPPL.2):17253–17260
Wikenius E, Moe V, Smith L, Heiervang ER, Berglund A (2019) DNA methylation changes in infants between 6 and 52 weeks. Sci Rep 9(1):1–12
Hannum G, Guinney J, Zhao L, Li Z, Hughes G, Sadda SV, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K (2013) Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49(2):359–367
Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P (2015) Reconfiguration of DNA methylation in aging, vol 151. Elsevier Ireland Ltd, Dublin
Jintaridth P, Mutirangura A (2010) Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics 41(2):194–200
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CPG island context. PLoS Genet. 2009;5(8)
Bormann F, Rodríguez-Paredes M, Hagemann S, Manchanda H, Kristof B, Gutekunst J, Raddatz G, Haas R, Terstegen L, Wenck H, Kaderali L, Winnefeld M, Lyko F (2016) Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell 15(3):563–571
Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A (2009) Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130(4):234–239
Bjornsson HT, Sigurdsson MI, Daniele Fallin M, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP (2008) Intra-individual change over time in DNA methylation with familial clustering. JAMA 299(24):2877–2883
Li X, Wang J, Wang L, Feng G, Li G, Yu M, Li Y, Liu C, Yuan X, Zang G, Li Z, Zhao L, Ouyang H, Quan Q, Wang G, Zhang C, Li O, Xiang J, Zhu J, Li W, Qi Z, Zhang K (2020) Impaired lipid metabolism by age-dependent DNA methylation alterations accelerates aging. Proc Natl Acad Sci U.S.A 117(8):4328–4336
Hadad N, Masser DR, Logan S, Wronowski B, Mangold CA, Clark N, Otalora L, Unnikrishnan A, Ford MM, Giles CB, Wren JD, Richardson A, Sonntag WE, Stanford DR, Freeman W (2016) Absence of genomic hypomethylation or regulation of cytosine-modifying enzymes with aging in male and female mice. Epigenetics Chromatin 9:30
Raddatz G, Hagemann S, Aran D, Söhle J, Kulkarni PP, Kaderali L, Hellman A, Winnefeld M, Lyko F (2013a) Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics Chromatin 6(1):36
Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10)
Weidner CI, Wagner W (2014) The epigenetic tracks of aging, vol 395. Walter de Gruyter GmbH, Berlin
Cedar H, Bergman Y (2012) Programming of DNA methylation patterns. Annu Rev Biochem 81(1):97–117
Jung M, Pfeifer GP (2015) Aging and DNA methylation. BMC Biol 13(1):7
Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, Gu H, Bock C, Meissner A, Göttgens B, Darlington GJ, Li W, Goodell MA (2014) Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14(5):673–688
Cole JJ, Robertson NA, Rather MI, Thomson JP, McBryan T, Sproul D, Wang T, Brock C, Clark W, Ideker T, Meehan RR, Miller RA, Brown-Borg HM, Adams PD (2017) Diverse interventions that extend mouse lifespan suppress shared age-associated epigenetic changes at critical gene regulatory regions. Genome Biol 18(1):58
Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner1 AP, Aviv1 A, Lohman K, Liu Y, Ferrucci L, Horvath S (2018) An epigenetic biomarker of aging for lifespan and healthspan. Aging 10(4):573–591
McEwen LM, O’Donnell KJ, McGill MG, Edgar RD, Jones M, MacIsaac JL, Lin DTS, Ramadori K, Morin A, Gladish N, Garg E, Unternaehrer E, Pokhvisneva I, Karnani N, Kee MZL, Klengel T, Adler NE, Barr RG, Letourneau N, Giesbrecht GF, Reynolds JN, Czamara D, Armstrong JM, Essex MJ, de Weerth C, Beijers R, Tollenaar MS, Bradley B, Jovanovic T, Ressler KJ, Steiner M, Entringer S, Wadhwa PD, Buss C, Bush NR, Binder EB, Thomas Boyce W, Meaney MJ, Horvath S, Kobor MS The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc Natl Acad Sci. 2019; 201820843
Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S (2019) DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11(2):303–327
Horvath S, Raj K (2018) DNA methylation-based biomarkers and the epigenetic clock theory of ageing, vol 19. Nature Publishing Group, Berlin
Sardo VL, Ferguson W, Erikson GA, Topol EJ, Baldwin KK, Torkamani A (2017) Influence of donor age on induced pluripotent stem cells. Nat Biotechnol 35(1):69–74
Raddatz Günter, Guzzardo PM, Olova N, Fantappié MR, Rampp M, Schaefer M, Reik W, Hannon G, Lyko F (2013b) Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci USA 110(21):8627–8631
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858
Li L, Zhuang Y, Zhao X, Li X (2019) Long non-coding RNA in neuronal development and neurological disorders, vol 10. Frontiers Media S.A., Lausanne
Cohen AL, Jia S (2014) Noncoding RNAs and the borders of heterochromatin. vol 5. NIH Public Access
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, RadhaDuttagupta EF, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guig R, Gingeras TR (2012) Landscape of transcription in human cells. Nature 489(7414):101–108
Wilusz JE, Sunwoo H, David L Spector long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23
Kato M, Chen X, Inukai S, Zhao H, Slack FJ (2011) Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17(10):1804–1820
Ibáñez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M (2006) Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5(3):235–246
Marcelo A, Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, MacOtela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR (2012) Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metabol 16(3):336–347
Lencastre AD, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ (2010) MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20(24):2159–2168
Zhang J, Liu Q, Zhang W, Li J, Li Z, Tang Z, Li Y, Han C, Hall SH, Zhang Y (2010) Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta biochimica et biophysica Sinica 42 (2)
Hooten NN, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE. 2010;5(5)
Li N, Bates DJ, An J, Terry DA, Wang E (2011) Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol Aging 32(5):944–955
Meier I, Fellini L, Jakovcevski M, Schachner M, Morellini F (2010) Expression of the snoRNA host gene gas5 in the hippocampus is upregulated by age and psychogenic stress and correlates with reduced novelty-induced behavior in C57BL/6 mice. Hippocampus 20(9):1027–1036
Trembinski JD, Bink DI, Theodorou K, Sommer J, Fischer A, van Bergen A, Kuo C-C, Costa IG, Schürmann C, Leisegang MS, Brandes RP, Alekseeva T, Brill B, Wietelmann A, Johnson CN, Spring-Connell A, Kaulich M, Werfel S, Engelhardt S, Hirt MN, Yorgan K, Eschenhagen T, Kirchhof L, Hofmann P, Jaé N., Wittig I, Hamdani N, Bischof C, Krishnan J, Houtkooper RH, Dimmeler S, Boon RA (2020) Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat Commun 11(1):2039
Nojima T, Tellier M, Foxwell J, de Almeida CR, Tan-Wong SM, Dhir S, Dujardin G, Dhir A, Murphy S, Proudfoot NJ (2018) Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-Loop formation, replication stress, and cellular senescence. Mol Cell 72(6):970–984
Sedivy JM, Kreiling JA, Neretti N, Cecco MD, Criscione SW, Hofmann JW, Zhao X, Ito T, Peterson AL (2013) Death by transposition - the enemy within? Bioessays 35 (12):1035–1043
Cecco MD, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, Le O, Beauséjour C, Ambati J, Ambati K, Simon M, Seluanov A, Gorbunova V, Slagboom EP, Helfand SL, Neretti N, Sedivy JM (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566(7742):73–78
Wang Z, Baulcombe DC (2020) Transposon age and non-CG methylation. Nat Commun 11(1):1–9
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, Levine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson J, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Hong ML, Dubois J, Yang H, Jun Y u, Wang J, Huang G, Jun G u, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Pia Abola A, Proctor MJ, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie RW, Bastide MDL, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen H, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JGR, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson SL, Jones TA, Kasif S, Kaspryzk A, Kennedy S, James Kent W, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit Arian F.A., Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh FR, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Patrinos A, Morgan MJ (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860– 921
Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Aabidine AZE, Pioger L, Nigumann P, Saccani S, Andrau JC, Gilbert N, Cristofari G (2019) The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell 74(3):555–570
Simon M, Meter MV, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, Cecco MD, Leonova KI, Kogan V, Helfand SL, Neretti N, Roichman A, Cohen HY, Meer M, Gladyshev VN, Antoch MP, Gudkov AV, Sedivy JM, Seluanov A, Gorbunova V (2019) LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 29(4):871–885
Gurdon JB The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. Development. 1962;10(4)
Velychko S, Adachi K, Kim K, Hou Y, MacCarthy CM, Guangming W u, Schöler HR (2019) Excluding Oct4 from Yamanaka cocktail unleashes the developmental potential of ipscs. Cell Stem Cell 25(6):737–753
Liu P, Chen M, Liu Y, Qi LS, Ding S (2018a) CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell 22(2):252–261
Buganim Y, Faddah DA, Jaenisch R (2013) Mechanisms and models of somatic cell reprogramming, vol 14. Nature Publishing Group, New York
Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, Oudenaarden AV, Jaenisch R (2012) Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150(6):1209–1222
Yang J, Rajan SS, Friedrich MJ, Lan G, Zou X, Ponstingl H, Garyfallos DA, Liu P, Bradley A, Metzakopian E (2019) Genome-scale CRISPRa screen identifies novel factors for cellular reprogramming. Stem Cell Rep 12(4):757–771
Wang F, Yin Y u, Ye X, Liu K, Zhu H, Wang L, Chiourea M, Okuka M, Ji G, Dan J, Zuo B, Li M, Zhang Q, Na L, Chen L, Pan X, Gagos S, Keefe DL, Liu L (2012) Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res 22(4):757–768
Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, Gui H u, Hsu SC, Shields EJ, Lyu X, Apostolou E, Hochedlinger K, Corces VG, Dekker J, Phillips-Cremins JE (2016) Local genome topology can exhibit an incompletely rewired 3D-folding state during somatic cell reprogramming. Cell Stem Cell 18(5):611–624
Churko JM, Lee J, Ameen M, Mingxia G u, Venkatasubramanian M, Diecke S, Sallam K, Im H, Wang G, Gold JD, Salomonis N, Snyder MP, Joseph C. W. u. (2017) Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. Nat Biomed Eng 1(10):826–837
Bartoccetti M, van der Veer BK, Luo X, Khoueiry R, She P, Bajaj M, Jiayi X u, Janiszewski A, Thienpont B, Pasque V, Koh KP (2020) Regulatory dynamics of Tet1 and Oct4 resolve stages of global DNA demethylation and transcriptomic changes in reprogramming. Cell Rep 30(7):2150–2169
Roost MS, Slieker RC, Bialecka M, Iperen LV, Gomes Fernandes MM, He N, Suchiman HED, Szuhai K, Carlotti F, De Koning EJP, Mummery CL, Heijmans BT, Lopes SM, De Sousa C (2017) DNAs methylation and transcriptional trajectories during human development and reprogramming of isogenic pluripotent stem cells. Nat Commun 8(1):1–11
Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K (2012) A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151(7):1617–1632
Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460(7259):1149–1153
Wang T, Hao W u, Li Y, Szulwach KE, Li L, Li X, Ping Chen I, Goldlust I., Chamberlain SJ, Dodd A, He G, Ananiev G, Ji WH, Yoon YS, Rudd KM, Miao Y u, Song CX, He C, Chang Q, Warren ST, Jin P (2013) Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat Cell Biol 15(6):700–711
Marión RM, de Silane IL, Mosteiro L, Gamache B, Abad M, Guerra C, Megías D, Serrano M, Blasco MA (2017) Common telomere changes during in vivo reprogramming and early stages of tumorigenesis. Stem Cell Rep 8(2):460–475
Manukyan M, Singh PB (2014) Epigenome rejuvination: HP1β mobility as a measure of pluripotent and senescent chromatin ground states. Sci Rep 4(1):1–8
Krijger PHL, Stefano BD, Wit ED, Limone F, Oevelen CV, Laat WD, Graf T (2016) Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell 18(5):597–610
Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J (2012) Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Reports 2(6):1579–1592
Growth-promoting E. R. (2013) Complete genome sequence of the sesbania symbiont and rice. Nucleic Acids Res 1(1256879):13–14
Watanabe A, Yamada Y, Yamanaka S (2013) Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier, vol 368. Royal Society, London
Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467(7313):285–290
Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R (2019) Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry 9(1):1–13
Papp B, Plath K (2013) Epigenetics of reprogramming to induced pluripotency, vol 152. Elsevier, Amsterdam
Sheng C, Jungverdorben J, Wiethoff H, Lin Q, Flitsch LJ, Eckert D, Hebisch M, Fischer J, Kesavan J, Weykopf B, Schneider L, Holtkamp D, Beck H, Till A, Wüllner U, Ziller MJ, Wagner W, Peitz M, Brüstle O (2018) A stably self-renewing adult blood-derived induced neural stem cell exhibiting patternability and epigenetic rejuvenation. Nat Commun 9(1):1–15
Olova N, Simpson DJ, Marioni RE, Chandra T (2019) Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell 18(1):e12877
Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai SI (2013) Sirt1 extends life span and delays aging in mice through the regulation of Nk2 Homeobox 1 in the DMH and LH. Cell Metab 18(3):416–430
Yang NC, Miao Lin H u (2005) The limitations and validities of senescence associated-β-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp Gerontol 40 (10):813–819
Gill D, Parry A, Santos F, Hernando-Herraez I, Stubbs TM, Milagre I, Reik W (2021) Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. bioRxiv, page 2021.01.15.426786
Göbel C., Goetzke R, Eggermann T, Wagner W (2018) Interrupted reprogramming into induced pluripotent stem cells does not rejuvenate human mesenchymal stromal cells. Sci Rep 8(1):1–7
Liu JY, Souroullas GP, Diekman BO, Krishnamurthy J, Hall BM, Sorrentino JA, Parker JS, Sessions GA, Gudkov AV, Sharpless NE (2019) Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proc Natl Acad Sci U.S.A 116 (7):2603– 2611
Guo L, Karoubi G, Duchesneau P, Shutova MV, Sung HK, Tonge P, Bear C, Rogers I, Nagy A, Waddell TK (2017) Generation of induced progenitor-like cells from mature epithelial cells using interrupted reprogramming. Stem Cell Rep 9(6):1780–1795
Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, Yang Jae-Hyun, Zhou S, Hoffmann EM, Karg MM, Schultz MB, Kane AE, Davidsohn N, Korobkina E, Chwalek K, Rajman LA, Church GM, Hochedlinger K, Gladyshev VN, Horvath S, Levine ME, Gregory-Ksander MS, Ksander BR, He Z, Sinclair DA (2020) Reprogramming to recover youthful epigenetic information and restore vision. Nature 588(7836):124–129
Mosteiro L, Pantoja C, Alcazar N, Marión RM, Chondronasiou D, Rovira M, Fernandez-Marcos PJ, Muñoz-Martin M, Blanco-Aparicio C, Pastor J, Gómez-López G, De Martino A, Blasco MA, Abad M, Serrano M (2016) Tissue damage and senescence provide critical signals for cellular reprogramming in vivo, vol 354
Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, Cherry ABC, Demirbas D, Tsankov AM, Zon LI, Rubin LL., Feinberg AP, Meissner A, Cowan CA, Daley GQ (2015) A comparison of non-integrating reprogramming methods. Nat Biotechnol 33(1):58–63
Mahmoudi S, Mancini E, Xu Lucy, Moore A, Jahanbani F, Hebestreit K, Srinivasan R, Li X, Devarajan K, Prélot L, Ang CE, Shibuya Y, Benayoun BA, Chang ALS, Wernig M, Wysocka J, Longaker MT, Snyder MP, Brunet A (2019) Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature 574(7779):553–558
Skamagki M, Correia C, Yeung P, Baslan T, Beck S, Zhang C, Ross CA, Dang L, Liu Z, Giunta S, Chang TP, Wang J, Ananthanarayanan A, Bohndorf M, Bosbach B, Adjaye J, Funabiki H, Kim J, Lowe S, Collins JJ, Lu CW, Li H, Zhao R, Kim K (2017) ZSCAN10 expression corrects the genomic instability of iPSCs from aged donors. Nat Cell Biol 19(9):1037–1048
Liu Y, Wu F, Zhang L, Wu X, Li D, Xin J, Xie J, Kong F, Wang W, Wu Q, Zhang D, Wang R, Gao S, Li W (2018b) Transcriptional defects and reprogramming barriers in somatic cell nucler reprogramming as revealed by single-embryo RNA sequencing. BMC Genom 19(1):734
Baumann V, Wiesbeck M, Breunig CT, Braun JM, Köferle A, Ninkovic J, Götz M, Stricker SH (2019) Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat Commun 10(1):1–12
Kang JG, Park JS, Ko JH, Kim YS (2019) Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci Rep 9(1):1–12
Kogut I, McCarthy SM, Pavlova M, Astling DP, Chen X, Jakimenko A, Jones KL, Getahun A, Cambier JC, Pasmooij AMG, Jonkman MF, Roop DR, Bilousova G (2018) High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat Commun 9 (1):1–15
Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, Manzanares M, Ortega S, Serrano M (2013) Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502(7471):358–345
Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, Soejima H, Moriwaki H, Yamanaka S, Woltjen K, Yamada Y (2014) Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156(4):663–677
Shibata H, Komura S, Yamada Y, Sankoda N, Tanaka A, Ukai T, Kabata M, Sakurai S, Kuze B, Woltjen K, Haga H, Ito Y, Kawaguchi Y, Yamamoto T, Yamada Y (2018) In vivo reprogramming drives Kras-induced cancer development. Nat Commun 9(1):1–16
Senís E, Mosteiro L, Wilkening S, Wiedtke E, Nowrouzi A, Afzal S, Fronza R, Landerer H, Abad M, Niopek D, Schmidt M, Serrano M, Grimm D (2018) AAVvector-mediated in vivo reprogramming into pluripotency. Nat Commun 9(1):1–14
Gore A, Li Z, Ho LF, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Je HL, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Belmonte JCI, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein Lawrence S.B., Zhang K (2011) Somatic coding mutations in human induced pluripotent stem cells. Nature 471(7336):63–67
Singovski G, Bernal C, Kuciak M, Siegl-Cachedenier I, Conod A, Altaba ARI In vivo epigenetic reprogramming of primary human colon cancer cells enhances metastases. J Mol Cell Biol. 2016;8(2)
Teng S, Li Y, Yang M, Qi R, Huang Y, Wang Q, Zhang Y, Chen S, Li S, Lin K, Cao Y, Ji Q, Qingyang G u, Cheng Y, Chang Z, Guo W, Wang P, Garcia-Bassets I, Lu ZJ, Wang D (2020) Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res 30(1):34–49
Son MJ, Son M-Y, Seol B, Kim M-J, Yoo CH, Han M-K, Cho YS (2013) Nicotinamide overcomes pluripotency deficits and reprogramming barriers. STEM CELLS 31(6):1121–1135
Li H, Collado M, Villasante A, Strati K, Ortega S, Cãamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460(7259):1136–1139
Tejas Y, Quivy JP, Almouzni G (2018) Chromatin plasticity: a versatile landscape that underlies cell fate and identity, vol 361. American Association for the Advancement of Science, Boston
Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, Waterfall JJ, Quivy JP, Almouzni G, Amigorena S (2018) The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359(6372):177–186
Li T, Ke Z, Tombline G, Macoretta N, Hayes K, Tian X, Lv R, Ablaeva J, Gilbert M, Bhanu NV, Yuan ZF, Garcia BA, Shi Y, Shi Y, Seluanov A, Gorbunova V (2017) Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Reports 9(5):1721– 1734
Cabreiro F, Catherine A u, Leung KY, Vergara-Irigaray N, Cochemé HM, Noori T, Weinkove D, Schuster E, Greene NDE, Gems D (2013) Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153(1):228–239
Chen J, Ou Y, Li Y, Hu S, Li WS, Liu Y Metformin extends C. elegans lifespan through lysosomal pathway. eLife. 2017;6
Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, Cabo RD (2013) Metformin improves healthspan and lifespan in mice. Nat Commun 4:2192
Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA (2016) Cell metabolism essay metformin as a tool to target aging
Kajbaf F, De Broe ME, Lalau JD (2016) Therapeutic concentrations of metformin: a systematic review, vol 55. Springer International Publishing, Berlin
Fang J, Yang J, Wu X, Zhang G, Li T, Wang X, Zhang H, Wang C, Liu GH, Wang L Metformin alleviates human cellular aging by upregulating the endoplasmic reticulum glutathione peroxidase 7. Aging Cell. 2018;17(4)
Vazquez-Martin A, Vellon L, Quirós PM, Cufí S, Galarreta ERD, Ferraros CO, Martin AG, Martin-Castillo B, Lopez-Otin C, Menendez JA (2012) Activation of AMP-activated protein kinase (AMPK) provides a metabolic barrier to reprogramming somatic cells into stem cells. Cell Cycle 11(5):974–989
Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, Woodward MA, Miller RA (2012) Rapamycin slows aging in mice. Aging Cell 11(4):675–682
Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11(1):35–46
Johnson SC, Rabinovitch PS, Kaeberlein M (2013) MTOR is a key modulator of ageing and age-related disease, vol 493. Nature Publishing Group, Berlin
Chen T, Li S, Yu J, Wan H, Ao G, Chen J, Long Y, Zhao J, Pei G (2011) Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell 10(5):908–911
He J, Kang L, Wu T, Zhang J, Wang H, Gao H, Yu Z, Bo H, Liu W, Kou Z, Zhang H, Gao S (2012) An elaborate regulation of mammalian target of rapamycin activity is required for somatic cell reprogramming induced by defined transcription factors. Stem Cells Dev 21 (14):2630–2641
Trepanier DJ, Gallant H, Legatt DF, Yatscoff RW (1998) Rapamycin: Distribution, pharmacokinetics and therapeutic range investigations: an update. In: Clinical Biochemistry, vol 31. Elsevier, pp 345–351
Fang Y, Westbrook R, Hill C, Boparai RK, Arum O, Spong A, Wang F, Javors MA, Chen J, Sun LY, Bartke A (2013) Duration of rapamycin treatment has differential effects on metabolism in mice. Cell Metab 17(3):456–462
Kaeberlein M (2014) Rapamycin and ageing when, for how long, and how much? J Genet Genomics 41(9):459–463
Bhullar KS, Hubbard BP (1852) Lifespan and healthspan extension by resveratrol. Elsevier, Amsterdam, p 2015
Ding DF, Li XF, Xu H, Wang Z, Liang QQ, Li CG, Wang Y (2013) Mechanism of resveratrol on the promotion of induced pluripotent stem cells. J Integr Med 11(6):389–396
Lee YL, Peng Q, Fong SW, Chen ACH, Lee KF, Ng EHY, Nagy A, Yeung WSB Sirtuin 1 facilitates generation of induced pluripotent stem cells from mouse embryonic fibroblasts through the miR-34a and p53 P. PLoS ONE. 2012;7(9)
Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Li LZ, Scherer B, Sinclair DA (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425(6954):191–196
Mukherjee S, Dudley JI, Das DK (2010) Dose-dependency of resveratrol in providing health benefits, vol 8. SAGE Publications, Singapore
Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J Biol Chem 277(47):45099–45107
Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S (2013) Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater 12(12):1154–1162
Weltner J, Balboa D, Katayama S, Bespalov M, Krjutškov K, Jouhilahti EM, Trokovic R, Kere J, Otonkoski T (2018) Human pluripotent reprogramming with CRISPR activators. Nat Commun 9(1):1–12
Fahy GM, Brooke RT, Watson JP, Good Z, Vasanawala SS, Maecker H, Leipold MD, Lin DTS, Kobor MS, Horvath S (2019) Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell 18(6):e13028
Horrington EM, Pope F, Lunsford W, McCay CM (1960) Age changes in the bones, blood pressure, and diseases of rats in parabiosis. Gerontology 4(1):21–31
Horvath S, Singh K, Raj K, Khairnar S, Sanghavi A, Shrivastava A, Zoller JA, Li CZ, Herenu CB, Canatelli-Mallat M, Lehmann M, Woods LC, Martinez AG, Wang T, Chiavellini P, Levine AJ, Chen H, Goya RG, Harold L, Katcher S (2020) Reversing age: dual species measurement of epigenetic age with a single clock. bioRxiv, page 2020.05.7.082917
Funding
RQ is grateful to the EPSRC Centre for Doctoral Training in Synthesis for Biology and Medicine (EP/L015838/1) for a studentship, generously supported by AstraZeneca, Diamond Light Source, Defence Science and Technology Laboratory, Evotec, GlaxoSmithKline, Janssen, Novartis, Pfizer, Syngenta, Takeda, UCB, and Vertex.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
de Lima Camillo, L.P., Quinlan, R.B.A. A ride through the epigenetic landscape: aging reversal by reprogramming. GeroScience 43, 463–485 (2021). https://doi.org/10.1007/s11357-021-00358-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-021-00358-6