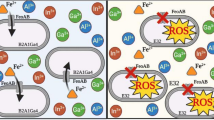
Abstract
To better understand the function of transporter in heavy metal detoxification of bacteria, the transporters associated with heavy metal detoxification in S. rhizophila JC1 were analyzed, among which four members were verified by RT-qPCR. In addition, the removal rates of four single metal ions (Cr6+, Cu2+, Zn2+, Pb2+) and polymetallic ions by strain JC1 were studied, respectively. We also researched the physiological response of strain JC1 to different metal stress via morphological observation, elemental composition, functional group and membrane permeability analysis. The results showed that in the single metal ion solution, removal capacities of Cu2+ (120 mg/L) and Cr6+ (80 mg/L) of S. rhizophila JC1 reached to 79.9% and 89.3%, respectively, while in polymetallic ions solution, the removal capacity of each metal ion all decreased, and in detail, the adsorption capacity was determined Cr6+>Cu2+>Zn2+>Pb2+ under the same condition. The physiological response analyses results showed that extracellular adsorption phenomena occurred, and the change of membrane permeability hindered the uptake of metal ions by bacteria. The analysis of transporters in strain JC1 genome illustrated that a total of 323 transporters were predicted. Among them, two, six and five proteins of the cation diffusion facilitator, resistance–nodulation–division efflux and P-type ATPase families were, respectively, predicted. The expression of corresponding genes showed that the synergistic action of correlative transporters played important roles in the process of adsorption. The comparative genomics analysis revealed that S. rhizophila JC1 has long-distance evolutionary relationships with other strains, but the efflux system of S. rhizophila JC1 contained the same types of metal transporters as other metal-resistant bacteria.
Graphical abstract







Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alimohamadi M, Abolhamd G, Keshtkar A (2005) Pb (II) and Cu (II) biosorption on Rhizopus arrhizus modeling mono-and multi-component systems. Miner Eng 18(13-14):1325–1330. https://doi.org/10.1016/j.mineng.2005.08.007
Amin A, Sarwar A, Saleem MA et al (2017) Expression and Purification of Transmembrane Protein MerE from Mercury-Resistant. J Microbiol Biotechnol 29:274–282. https://doi.org/10.4014/jmb.1704.04062
Anton A, Weltrowski A, Haney CJ, Franke S, Grass G, Rensing C, Nies DH (2004) Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans CH34 and Escherichia coli. J Bacteriol 186(22):7499–7507. https://doi.org/10.1128/JB.186.22.7499-7507.2004
Arguello JM, Manuel GG, Raimunda D (2011) Bacterial transition metal P1B-ATPases: transport mechanism and roles in virulence. Biochemistry 50(46):9940–9949. https://doi.org/10.1016/B978-0-12-307212-2.50008-9
Aryal M, Ziagova MG, Liakopoulou-Kyriakides M (2012) Cu (II) biosorption and competitive studies in multi-ions aqueous systems by Arthrobacter sp. Sphe3 and Bacillus sphaericus cells: Equillibrium and thermodynamic studies. Water Air Soil 23(8):5119–5130. https://doi.org/10.1007/s11270-012-1263-9
Barber ZS, Uebe R, Davidov GY et al (2016) Disease-Homologous Mutation in the Cation Diffusion Facilitator Protein MamM Causes Single-Domain Structural Loss and Signifies Its Importance. Nature 6:31933. https://doi.org/10.1038/srep31933
Borremans B, Hobman JL, Provoost A, Brown NL et al (2001) Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol 183:5651–5658. https://doi.org/10.1128/jb.183.19.5651-5658.2001
Bruins MR, Kapil S, Oehme FW (2000) Microbial resistance to metals in the environment. Ecotoxicol Environ Saf 45(3):198–207. https://doi.org/10.1006/eesa.1999.1860
Chen S, Li X, Wang Y et al (2018) Induction of Escherichia coli into a VBNC state through chlorination/chloramination and differences in characteristics of the bacterium between states. Water Res 142:279–288. https://doi.org/10.1016/j.watres.2018.05.055
David K, Michael M (2002) Structure, properties and regulation of magnesium transport proteins. Biometals 15(3):261–270. https://doi.org/10.1023/A:1016078832697
Diels L, Dong QH et al (1995) The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol Biotechnol 14(2):142–153. https://doi.org/10.1007/BF01569896
El-Baghdady KZ, El-Shatoury EH, Abdullah OM et al (2018) Biogenic production of silver nanoparticles by Enterobacter cloacae Ism26. Turk J Biol 42(4):319–321. https://doi.org/10.3906/biy-1801-53
Fathima A, Rao JR (2017) Is Cr(III) toxic to bacteria: toxicity studies using Bacillus subtilis and Escherichia coli as model organism. Arch Microbiol 200(3):453–462. https://doi.org/10.1007/s00203-017-1444-4
Franke S, Grass G, Rensing C, Nies DH (2003) Molecular analysis of the copper-transporting CusCFBA efflux system from Escherichia coli. J Bacteriol 185:3804–3812. https://doi.org/10.1128/JB.185.13.3804-3812.2003
Gola D, Chauhan N, Malik A, Shaikh ZA (2017) Bioremediation Approach for Handling. Taylor and Francis, New York
Grass G, Rensing C (2001) CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286:902–908. https://doi.org/10.1006/bbrc.2001.5474
Grass G, Grosse C, Nies DH (2000) Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J Bacteriol 182(5):1390–1398. https://doi.org/10.1128/JB.182.5.1390-1398.2000
Grass G, Thakali K, Klebba PE et al (2004) Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J Bacteriol 186:5826–5833. https://doi.org/10.1128/JB.186.17.5826-5833.2004
Große C, Anton A, Hoffmann T, Franke S, Schleuder G, Nies DH (2004) Identifification of a regulatory pathway that controls the heavy metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch Microbiol 182:109–118. https://doi.org/10.1007/s00203-004-0670-8
Guskov A, Eshaghi S (2012) The mechanisms of Mg2+ and Co2+ transport by the cora family of divalent cation transporters. Curr Top Membr 69:393–414. https://doi.org/10.1016/B978-0-12-394390-3.00014-8
Higuchi T, Hattori M, Tanaka Y, Ishitani R, Nureki O (2009) Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein. Proteins 76(3):768–771. https://doi.org/10.1002/prot.22444
Hiraizumi M, Yamashita K, Nishizawa T, Nureki O (2019) Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365:1149–1155. https://doi.org/10.1101/666321
Hložková K, Suman J, Strnad H et al (2013) Characterization of pbt genes conferring increased Pb2+ and Cd2+ tolerance upon Achromobacter xylosoxidans A8. Res Microbiol 164(10):1009–1018. https://doi.org/10.1016/j.resmic.2013.10.002
Howell SC, Mesleh MF, Opella SJ (2005) NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry 44:5196–5206. https://doi.org/10.1021/bi048095v
Hu N, Zhao B (2007) Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol Lett 267(1):17–22. https://doi.org/10.1111/j.1574-6968.2006.00505.x
Hwang H, Hazel A, Lian P et al (2019) A Minimal Membrane Metal Transport System: Dynamics and Energetics of mer Proteins. J Comput Chem 41(6). https://doi.org/10.1002/jcc.26098
Jin HP, Chon HT (2016) Characterization of cadmium biosorption by Exiguobacterium sp. isolated from farmland soil near Cu-Pb-Zn mine. Environ Sci Pollut Res 23(12):11814–11822. https://doi.org/10.1007/s11356-016-6335-8
Jurkiewicz P, Olżyńska A, Cwiklik L, Conte E et al (2012) Biophysics of lipid bilayers containing oxidatively modified phospholipids: insights from fluorescence and EPR experiments and from MD simulations. Biochim Biophys Acta 1818(10):2388–2402. https://doi.org/10.1016/j.bbamem.2012.05.020
Kambe T (2012) Molecular architecture and function of ZnT transporters. Curr Top Membr 69:199–220. https://doi.org/10.1016/B978-0-12-394390-3.00008-2
Kang CH (2016) Heavy metal and antibiotic resistance of ureolytic bacteria and their immobilization of heavy metals. Ecol Eng 97:304–312. https://doi.org/10.1016/j.ecoleng.2016.10.016
Kim EH, Nies DH, McEvoy MM, Rensing C (2011) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 10(193):2381–2387. https://doi.org/10.1128/JB.01323-10
Klein J, Lewinson O (2011) Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metall Int Bio Sci 3(11):1098–1108. https://doi.org/10.1039/c1mt00073j
Knoop V, Groth-Malonek M, Gebert M et al (2005) Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol Gen Genomics 274(3):205–216. https://doi.org/10.1007/s00438-005-0011-x
Liang JQ, Zhang MZ, Lu MM, Li ZF et al (2016) Functional characterization of a csoR-cueA divergon in Bradyrhizobium liaoningense CCNWSX0360, involved in copper, zinc and cadmium cotolerance. Sci Rep-UK 6:35155. https://doi.org/10.1038/srep3515
Liu H, Du Y, Wang X, Sun L (2004) Chitosan kills bacteria through cell membrane damage. Int J Food Microbiol 95(2):147–155. https://doi.org/10.1016/j.ijfoodmicro.2004.01.022
Loh B, Grant C, Hancock RE (1984) Use of the fluorescent Probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa, Antimicrob. Agents Chemother 26(4):546–551. https://doi.org/10.1128/AAC.26.4.546
Lu M, Fu D (2007) Structure of the zinc transporter YiiP. Science 317:1746–1748. https://doi.org/10.1126/science.1143748
Luo S, Li X, Chen L, Chen J et al (2014) Layer-by-layer strategy for adsorption capacity fattening of endophytic bacterial biomass for highly effective removal of heavy metals. Chem Eng J 239:312–321. https://doi.org/10.1016/j.cej.2013.11.029
Lv Y, Zhang Y, Song J, Xu G (2020) Regulation of the activity and kinetic properties of alkaline phosphatase in eutrophic water bodies by phosphorus and heavy metals. Desalin Water Treat 177:131–138. https://doi.org/10.5004/dwt.2020.24924
Ma Z, Jacobsen FE, Giedroc DP (2009) Coordination chemistry of bacterial metal transport and sensing. Chem Rev 109(10):4644–4681. https://doi.org/10.1021/cr900077w
Maseda H, Kitao M, Shima E, Yoshihara E, Nakae T (2002) A novel assembly process of the multicomponent xenobiotic efflux pump in Pseudomonas aeruginosa. Mol Microbiol 3(46):677–686. https://doi.org/10.1046/j.1365-2958.2002.03197.x
Masood F, Malik A (2015) Single and multi-component adsorption of metal ions by Acinetobacter sp. FM4. Sep Sci Technol 50(6):892–900. https://doi.org/10.1080/01496395.2014.969378
Meng D, Bruschweiler LL, Zhang F, Brüschweiler R (2015) Modulation and Functional Role of the Orientations of the N- and P-Domains of Cu+ -Transporting ATPase along the Ion Transport Cycle. Biochemistry 54:5095–5102. https://doi.org/10.1021/acs.biochem.5b00420
Mertens J, Degryse F, Springael D, Smolders E (2007) Zinc toxicity to nitrification in soil and soilless culture can be predicted with the same biotic ligand model. Environ Sci Technol 41(8):2992–2997. https://doi.org/10.1021/es061995+
Mohite BV, Koli SH, Patil SV (2018) Heavy Metal Stress and Its Consequences on Exopolysaccharide (EPS)-Producing Pantoea agglomeran. Appl Biochem Biotechnol 186(1):199–216. https://doi.org/10.1007/s12010-018-2727-1
Moraleda MA, Pérez J, Extremera AL, Dorado JM (2010) Differential regulation of six heavy metal efflux systems in the response of Myxococcus xanthus to copper. Appl Biochem Biotechnol 76(18):6069–6076. https://doi.org/10.1128/AEM.00753-10
Murĺnová S, Dercová K (2014) Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int J Microbiol 2014:873081–873081. https://doi.org/10.1155/2014/873081
Nagakubo S, Nishino K, Hirata T, Yamaguchi A (2002) The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 15(184):4161–4167. https://doi.org/10.1128/JB.184.15.4161-4167.2002
Nies DH (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27(2):313–339. https://doi.org/10.1016/S0168-6445%2803%2900048-2
Nies DH, Rehbein G, Hoffmann T, Baumann C, Grosse C (2006) Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J Mol Microbiol Biotechnol 11(1-2):82–93. https://doi.org/10.1159/000092820
Novinscak A, Filion M (2011) Effect of soil clay content on RNA isolation and on detection and quantification of bacterial gene transcripts in soil by quantitative reverse transcription-PCR. Appl Environ Microbiol 77:6249–6252. https://doi.org/10.1128/AEM.00055-11
Osman D, Cavet JS (2011) Metal sensing in Salmonella: implications for pathogenesis. Elsevier Science & Technology, London
Pal A, Bhattacharjee S, Saha J et al (2021) Bacterial survival strategies and responses under heavy metal stress: a comprehensive overview. Crit Rev Microbiol. https://doi.org/10.1080/1040841X.2021.1970512
Panja S, Aich P, Jana B, Basu T (2008) How does plasmid DNA penetrate cell membranes in artificial transformation process of Escherichia coli? Mol Membr Biol 25(5):411–422. https://doi.org/10.1016/S0140-6736(00)04817-0
Paulsen IT, Saier MH (1997) A novel family of ubiquitous heavy metal ion transport proteins. J Membrane Boil 156:99–103. https://doi.org/10.1007/s002329900192
Pontel LB, Audero ME, Espariz M, Checa SK, Soncini FC (2010) GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Oral Microbiol 66(3):814–825. https://doi.org/10.1111/j.1365-2958.2007.05963.x
Prabhakaran P, Ashraf MA, Aqma WS (2016) Microbial stress response to heavy metals in the environment. RSC Adv 6(111):109862–109877. https://doi.org/10.1039/C6RA10966G
Rensing C, Pribyl T, Nies DH (1997) New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacter 179(22):6871. https://doi.org/10.1128/jb.179.22.6871-6879.1997
Rozycki TV, Nies DH (2009) Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96(2):115–139. https://doi.org/10.1007/s10482-008-9284-5
Saier MH, Tam R, Rerzer A (2010) Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol 11(5):841–847. https://doi.org/10.1111/j.1365-2958.1994.tb00362.x
Sasaki Y, Minakawa T, Miyazaki A et al (2005) Functional dissection of a mercuric ion transporter, MerC, from Acidithiobacillus ferrooxidans. Biosci Biotechnol Biochem 69:1394–1402. https://doi.org/10.1271/bbb.69.1394
Schlesinger MJ, Levinthal C (1989) Molecular Biology. Elsevier, London, pp 210–222
Schmidt CS, Alavi M, Cardinale M et al (2012) Stenotrophomonas rhizophila DSM14405T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol Fertil Soils 48(8):947–960. https://doi.org/10.1007/s00374-012-0688-z
Schué M, Dover LG, Besra GS et al (2009) Sequence and analysis of a plasmid-encoded mercury resistance operon from Mycobacterium marinum identifies MerH, a new mercuric ion transporter. J Bacteriol 191:439–444. https://doi.org/10.1128/JB.01063-08
Selvaraj U, Venu-Babu P, Thilagaraj WR (2018) Application of H412R mutant alkaline phosphatase for removal of heavy metals from single-ion solutions and effluents. Int J Environ Sci Technol 1-8. https://doi.org/10.1007/s13762-018-1730-y
Silver S, Phung LT (1996) Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 50:753–789. https://doi.org/10.1021/acs.biochem.6b01022
Smith AT, Ross MO, Hoffman BM, Rosenzweig AC (2017) Metal Selectivity of a Cd-, Co-, and Zn-Transporting P1B-type ATPase. Biochemistry 56(1):85–95. https://doi.org/10.1016/j.jmb.2019.12.025
Song DG, Li XX, Cheng YZ, Xiao X et al (2017) Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci Rep 7(1):3239. https://doi.org/10.1038/s41598-017-03558-3
Sun SC, Chen JX, Wang YG, Leng FF, Zhao J, Chen K (2021) Molecular mechanisms of heavy metals resistance of Stenotrophomonas rhizophila JC1 by whole genome sequencing. Arch Microbiol. https://doi.org/10.1007/S00203-021-02271-0
Taghavi S, Lesaulnier C, Monchy S, Wattiez R et al (2009) Lead (II) resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Antonie Van Leeuwenhoek 96:171–182. https://doi.org/10.1007/s10482-008-9289-0
Tam HK, Malviya VN, Foong WE, Herrmann A et al (2019) Binding and transport of carboxylated drugs by the multidrug transporter AcrB. J Mol Biol 432(4):861–877. https://doi.org/10.1186/1471-2180-13-79
Valencia EY, Braz VS, Guzzo C, Marques MV (2013) Two RND proteins involved in heavy metal efflux in Caulobacter crescentus belong to separate clusters within proteobacteria. BMC Microbiol 13(1):79–79. https://doi.org/10.1016/j.jconrel.2006.09.071
Vijayaraghavan K, Balasubramanian R (2015) Is biosorption suitable for decontamination of metal-bearing wastewaters? A critical review on the state-of-the-art of biosorption processes and future directions. J Environ Manag 160:283–296. https://doi.org/10.1016/j.jenvman.2015.06.030
Wang JP, Li GY, Yin HL, An TC (2020a) Bacterial response mechanism during biofilm growth on different metal material substrates: EPS characteristics, oxidative stress and molecular regulatory network analysis. Environ Res 185:109451. https://doi.org/10.1016/j.envres.2020.109451
Wang QF, Li Q, Lin Y, Hou Y et al (2020b) Biochemical and genetic basis of cadmium biosorption by Enterobacter ludwigii LY6, isolated from industrial contaminated soil. Environ Pollut 264:114637. https://doi.org/10.1016/j.envpol.2020.114637
Wang YG, Sun SC et al (2021) Optimization and mechanism exploration for Escherichia coli transformed with plasmid pUC19 by the combination with ultrasound treatment and chemical method. Ultrason Sonochem. https://doi.org/10.1016/j.ultsonch.2021.105552
Waschuk SA, Elton EA, Darabie AA, Fraser PE, McLaurin JA (2001) Cellular membrane composition defines A beta-lipid interactions. J Biol Chem 276:33561–33568. https://doi.org/10.1002/mbo3.182
Xiao XY, Wang MW, Zhu HW et al (2017) Response of soil microbial activities and microbial community structure to vanadium stress. Ecotoxicol Environ Saf 142:200–206. https://doi.org/10.1016/j.ecoenv.2017.03.047
Yang L, Lu S, Belardinelli J, Claustre EH, Jones V et al (2014) RND transporters protect Corynebacterium glutamicum from antibiotics by assembling the outer membrane. Microbiologyopen 3(4):484–496. https://doi.org/10.1016/j.carbpol.2013.09.021
Yang JS, Yang FL, Yang Y, Xing GL et al (2016) A proposal of“core enzyme” bioindicator in long-term Pb-Zn ore pollution areas based on topsoil property analysis. Environ Pollut 213:760–769. https://doi.org/10.1016/j.envpol.2016.03.030
Ye SH, Zhang MP, Yang H et al (2014) Biosorption of Cu2+, Pb2+ and Cr6+ by a novel exopolysaccharide from Arthrobacter ps-5. Carbohydr Polym 101:50–56. https://doi.org/10.1016/j.carbpol.2013.09.021
Zhang SH, Ye CS, Lin HR, Lv L, Yu X (2015) UV Disinfection induces a VBNC state in Escherichia coli and Pseudomonas aeruginosa. Environ Sci Technol 49(3):1721–1728. https://doi.org/10.1021/es505211
Zhang ZL, Cai RH, Zhang WH, Fu YN, Jiao NZ (2017) A Novel Exopolysaccharide with Metal Adsorption Capacity Produced by a Marine Bacterium Alteromonas sp. JL2810. Mar Drugs. https://doi.org/10.3390/md15
Authors, contributions
Jixiang Chen and Yonggang Wang designed the study; Shangchen Sun, Kexin Zhang and Yamiao Wu performed the study; Shangchen Sun, Yonggang Wang, Feifan Leng and Ning Zhu analyzed the data; Shangchen Sun and Kexin Zhang done writing—original draft preparation:; Jixiang Chen and Feifan Leng done writing-review and editing:; Jixiang Chen and Yonggang Wang contributed to funding acquisition. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31760028) and (No. 32160066).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Responsible Editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, S., Zhang, K., Wu, Y. et al. Transporter drives the biosorption of heavy metals by Stenotrophomonas rhizophila JC1. Environ Sci Pollut Res 29, 45380–45395 (2022). https://doi.org/10.1007/s11356-022-18900-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-18900-0