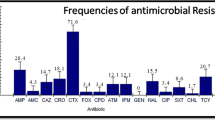
Abstract
The rapid development of antibiotic-resistant bacteria (ARB) has been of concern worldwide. In this study, antibiotic resistance genes (ARGs) were investigated in antibiotic-resistant Escherichia coli isolated from surface water samples (rivers, n = 17; Taihu Lake, n = 16) and from human, chicken, swine, and Egretta garzetta sources in the Taihu Basin. E. coli showing resistance to at least five drugs occurred in 31, 67, 58, 27, and 18 % of the isolates from surface water (n = 665), chicken (n = 27), swine (n = 29), human (n = 45), and E. garzetta (n = 15) sources, respectively. The mean multi-antibiotic resistance (MAR) index of surface water samples (0.44) was lower than that of chicken (0.64) and swine (0.57) sources but higher than that of human (0.30) and E. garzetta sources (0.15). Ten tetracycline, four sulfonamide, four quinolone, five β-lactamase, and two streptomycin resistance genes were detected in the corresponding antibiotic-resistant isolates. Most antibiotic-resistant E. coli harbored at least two similar functional ARGs. Int-I was detected in at least 57 % of MAR E. coli isolates. The results of multiple correspondence analysis and Spearman correlation analysis suggest that antibiotic-resistant E. coli in water samples were mainly originated from swine, chicken, and/or human sources. Most of the ARGs detected in E. garzetta sources were prevalent in other sources. These data indicated that human activities may have contributed to the spread of ARB in the aquatic environment.



Similar content being viewed by others
References
Abhirosh C, Sherin V, Thomas AP, Hatha AAM, Mazumder A (2011) Potential public health significance of faecal contamination and multidrug-resistant Escherichia coli and Salmonella serotypes in a lake in India. Public Health 125:377–379
Akiyama T, Savin MC (2010) Populations of antibiotic-resistant coliform bacteria change rapidly in a wastewater effluent dominated stream. Sci Total Environ 408:6192–6201
Alali WQ, Scott HM, Harvey RB, Norby B, Lawhorn DB, Pillai SD (2008) Longitudinal study of antimicrobial resistance among Escherichia coli isolates from integrated multisite cohorts of humans and swine. Appl Environ Microbiol 74:3672–3681
Allen SE, Boerlin P, Janecko N, Lumsden JS, Barker IK, Pearl DL, Reid-Smith RJ, Jardine C (2011) Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl Environ Microbiol 77:882–888
Baquero F, Martinez JL, Canton R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Ben-Mahrez K, Sioud M (2010) Presence of STRA-STRB linked streptomycin-resistance genes in clinical isolate of Escherichia coli 2418. Arch Inst Pasteur Tunis 87:77–81
Chen H, Shu W, Chang X, J-a C, Guo Y, Tan Y (2010) The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ Pollut 158:2459–2464
Chen B, Zheng W, Yu Y, Huang W, Zheng S, Zhang Y, Guan X, Zhuang Y, Chen N, Topp E (2011) Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River, Fujian Province, China. Appl Environ Microbiol 77:148–155
Cummings DE, Archer KF, Arriola DJ, Baker PA, Faucett KG, Laroya JB, Pfeil KL, Ryan CR, Ryan KRU, Zuill DE (2011) Broad dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ Sci Technol 45:447–454
Henriques IS, Fonseca F, Alves A, Saavedra MJ, Correia A (2006) Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res Microbiol 157:938–947
Hu J, Shi J, Chang H, Li D, Yang M, Kamagata Y (2008) Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ Sci Technol 42:3415–3420
Hussein AIA, Ahmed AM, Sato M, Shimamoto T (2009) Characterization of integrons and antimicrobial resistance genes in clinical isolates of Gram-negative bacteria from Palestinian hospitals. Microbiol Immunol 53:595–602
Ibekwe AM, Murinda SE, Graves AK (2011) Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. Plos One 6(6):e20819
Kich JD, Coldebella A, Mores N, Nogueira MG, Cardoso M, Fratamico PM, Call JE, Fedorka-Cray P, Luchansky JB (2011) Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina, Brazil. Int J Food Microbiol 151:307–313
Kiiru J, Kariuki S, Goddeeris BM, Butaye P (2012) Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol 12:155
Knapp CW, Dolfing J, Ehlert PAI, Graham DW (2010) Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol 44:580–587
Koljalg S, Truusalu K, Vainumaee I, Stsepetova J, Sepp E, Mikelsaar M (2009) Persistence of Escherichia coli clones and phenotypic and denotypic antibiotic resistance in recurrent urinary tract infections in childhood. J Clin Microbiol 47:99–105
Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C (2009) Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol 75:559–566
Levesque C, Piche L, Larose C, Roy PH (1995) PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39:185–191
Li X, Watanabe N, Xiao C, Harter T, McCowan B, Liu Y, Atwill ER (2014) Antibiotic-resistant E. coli in surface water and groundwater in dairy operations in Northern California. Environ Monit Assess 186:1253–1260
Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lue D, Huang L, Zhang Y, Liu J, Wang M (2009) High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac (6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother 53:519–524
Magalhaes VD, Schuman W, Castilho BA (1998) A new tetracycline resistance determinant cloned from Proteus mirabilis. Biochim Biophys Acta Gene Struct Expr 1443:262–266
Martinez JL (2008) Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367
Momtaz H, Rahimi E, Moshkelani S (2012) Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet Med 57:193–197
Nagachinta S, Chen J (2008) Transfer of class 1 integron-mediated antibiotic resistance genes from Shiga toxin-producing Escherichia coli to a susceptible E. coli K-12 strain in storm water and bovine feces. Appl Environ Microbiol 74:5063–5067
Olaniran AO, Naicker K, Pillay B (2009) Antibiotic resistance profiles of Escherichia coli isolates from river sources in Durban, South Africa. World J Microbiol Biotechnol 25:1743–1749
Ram S, Vajpayee P, Shanker R (2007) Prevalence of multi-antimicrobial-agent resistant, Shiga toxin and enterotoxin producing Escherichia coli in surface waters of river Ganga. Environ Sci Technol 41:7383–7388
Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC (2006) qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 50:2872–2874
Rodriguez-Martinez JM, Eliecer Cano M, Velasco C, Martinez-Martinez L, Pascual A (2011) Plasmid-mediated quinolone resistance: an update. J Infect Chemother 17:149–182
Sayah RS, Kaneene JB, Johnson Y, Miller R (2005) Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol 71:1394–1404
Sharma J, Sharma M, Ray P (2010) Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res 132:332–336
Sidrach-Cardona R, Hijosa-Valsero M, Marti E, Balcazar JL, Becares E (2014) Prevalence of antibiotic-resistant fecal bacteria in a river impacted by both an antibiotic production plant and urban treated discharges. Sci Total Environ 488:220–227
Soufi L, Abbassi MS, Saenz Y, Vinue L, Somalo S, Zarazaga M, Abbas A, Dbaya R, Khanfir L, Ben Hassen A, Hammami S, Torres C (2009) Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog Dis 6:1067–1073
Storteboom H, Arabi M, Davis JG, Crimi B, Pruden A (2010) Tracking antibiotic resistance genes in the South Platte River basin using molecular signatures of urban, agricultural, and pristine sources. Environ Sci Technol 44:7397–7404
Su H-C, Ying G-G, Tao R, Zhang R-Q, Zhao J-L, Liu Y-S (2012) Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ Pollut 169:42–49
Sunde M, Norstrom M (2005) The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemother 56:87–90
Tacão M1, Moura A, Correia A, Henriques I (2014) Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res 48:100–107
Tamminen M, Karkman A, Lohmus A, Muziasari WI, Takasu H, Wada S, Suzuki S, Virta M (2011) Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ Sci Technol 45:386–391
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang HH, Schaffner DW (2011) Antibiotic resistance: how much do we know and where do we go from here? Appl Environ Microbiol 77:7093–7095
Wang C, Gu XC, Zhang SH, Wang PF, Guo C, Gu J, Hou J (2013) Characterization of antibiotic-resistance genes in antibiotic resistance Escherichia coli isolates from a lake. Arch Environ Contam Toxicol 65:635–641
Woo PCY, To APC, Lau SKP, Yuen KY (2003) Facilitation of horizontal transfer of antimicrobial resistance by transformation of antibiotic-induced cell-wall-deficient bacteria. Med Hypotheses 61:503–508
Zhang T, Zhang M, Zhang X, Fang HH (2009a) Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environ Sci Technol 43:3455–3460
Zhang X, Wu B, Zhang Y, Zhang T, Yang L, Fang HHP, Ford T, Cheng S (2009b) Class 1 integronase gene and tetracycline resistance genes tetA and tetC in different water environments of Jiangsu Province, China. Ecotoxicology 18:652–660
Zhang XY, Ding LJ, Yue J (2009c) Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in northeastern China. Microb Drug Resist 15:323–328
Zhang YL, Marrs CF, Simon C, Xi CW (2009d) Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci Total Environ 407:3702–3706
Zhang W-J, Xu X-R, Schwarz S, Wang X-M, Dai L, Zheng H-J, Liu S (2014) Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J Antimicrob Chemother 69:385–389
Zhao L, Chen X, Zhu X, Yang W, Dong L, Xu X, Gao S, Liu X (2009) Prevalence of virulence factors and antimicrobial resistance of uropathogenic Escherichia coli in Jiangsu province (China). Urology 74:702–707
Acknowledgments
This work was supported by the National Foundation for the Natural Science Foundation of JiangSu Province (BK2012413), Innovative Research Groups (NO. 51421006), National Natural Science Foundation of China (NO. 51379063), and for Distinguished Young Scholars (NO. 51225901) and Research Fund for innovation team of Ministry of Education (IRT13061).
Compliance with ethical standards
There are no potential conflicts of interest. All the data in this manuscript have not been published previously (partly or in full). All the authors agreed with this submission.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Zhang, S.H., Lv, X., Han, B. et al. Prevalence of antibiotic resistance genes in antibiotic-resistant Escherichia coli isolates in surface water of Taihu Lake Basin, China. Environ Sci Pollut Res 22, 11412–11421 (2015). https://doi.org/10.1007/s11356-015-4371-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4371-4