Abstract
Monitoring of marine microalgae is important to predict and manage harmful algae blooms. It currently relies mainly on light-microscopic identification and enumeration of algal cells, yet several molecular tools are currently being developed to complement traditional methods. MIcroarray Detection of Toxic ALgae (MIDTAL) is an FP7-funded EU project aiming to establish a hierarchical multispecies microarray as one of these tools. Prototype arrays are currently being tested with field samples, yet the analysis of the large quantities of data generated by these arrays presents a challenge as suitable analysis tools or protocols are scarce. This paper proposes a two-part protocol for the analysis of the MIDTAL and other hierarchical multispecies arrays: Signal-to-noise ratios can be used to determine the presence or absence of signals and to identify potential false-positives considering parallel and hierarchical probes. In addition, normalized total signal intensities are recommended for comparisons between microarrays and in order to relate signals for specific probes to cell concentrations using external calibration curves. Hybridization- and probe-specific detection limits can be calculated to help evaluate negative results. The suggested analyses were implemented in “GPR-Analyzer”, a platform-independent and graphical user interface-based application, enabling non-specialist users to quickly and quantitatively analyze hierarchical multispecies microarrays. It is available online at http://folk.uio.no/edvardse/gpranalyzer.




Similar content being viewed by others
References
Ahn S, Kulis DM, Erdner DL, Anderson DM, Walt DR (2006) Fiber-optic microarray for simultaneous detection of multiple harmful algal bloom species. App Env Microbiol 72:5742–5749. doi:10.1111/j.1550-7408.2000.tb00014.x
Ayers K, Rhodes LL, Tyrrell J, Gladstone M, Scholin C (2005) International accreditation of sandwich hybridisation assay format DNA probes for microalgae. N Z J Mar Freshwat Res 39:1225–1231. doi:10.1080/00288330.2005.9517388
Behnke A, Engel M, Christen R, Nebel M, Klein RR, Stoeck T (2011) Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Env Microbiol 13:340–349. doi:10.1111/j.1462-2920.2010.02332.x
Costas BA, McManus G, Doherty M, Katz LA (2007) Use of species-specific primers and PCR to measure the distributions of planktonic ciliates in coastal waters. Limnol Oceanogr Meth 5:163–173. doi:10.4319/lom.2007.5.163
Diercks S, Metfies K, Medlin LK (2008) Molecular probe sets for the detection of toxic algae for use in sandwich hybridization formats. J Plankton Res 30:439–448. doi:10.1093/plankt/fbn009
Dittami SM, Edvardsen B (2012) Culture conditions influence cellular RNA content in ichthyotoxic flagellates of the genus Pseudochattonella (Dictyochophyceae). J Phycol in press. doi:10.1111/j.1529-8817.2012.01183.x
Eller G, Tobe K, Medlin LK (2007) Hierarchical probes at various taxonomic levels in the Haptophyta and a new division level probe for the Heterokonta. J Plankton Res 29:629–640. doi:10.1093/plankt/fbm045
Galluzzi L, Penna A, Bertozzini E, Vila M, Garces E, Magnani M (2004) Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). App Env Microbiol 70:1199–1206. doi:10.1128/AEM.70.2.1199-1206.2004
Gescher C, Metfies K, Medlin LK (2008) The ALEX CHIP—development of a DNA chip for identification and monitoring of Alexandrium. Harmful Algae 7:485–494. doi:10.1016/j.hal.2007.11.001
Groben R, John U, Eller G, Lange M, Medlin LK (2004) Using fluorescently-labelled rRNA probes for hierarchical estimation of phytoplankton diversity—a mini-review. Nova Hedwigia 79:313–320. doi:10.1127/0029-5035/2004/0079-0313
Humbert JF, Quiblier C, Gugger M (2010) Molecular approaches for monitoring potentially toxic marine and freshwater phytoplankton species. Analyt Bioanalyt Chem 397:1723–1732. doi:10.1007/s00216-010-3642-7
Huyghe A, Francois P, Charbonnier Y, Tangomo-Bento M, Bonetti E-J, Paster BJ, Bolivar I, Baratti-Mayer D, Pittet D, Schrenzel J (2008) Novel microarray design strategy to study complex bacterial communities. Appl Environ Microbiol 74:1876–1885. doi:10.1128/AEM.01722-07
John U (2004) Development of specific rRNA probes to distinguish between geographic clades of the Alexandrium tamarense species complex. J Plankton Res 27:199–204. doi:10.1093/plankt/fbh160
Lange M, Guillou L, Vaulot D, Simon N, Amann RI, Ludwig W, Medlin LK (1996) Identification of the class Prymnesiophyceae and the genus Phaeocystis with ribosomal RNA-targeted nucleic acid probes detected by flow cytometry. J Phycol 32:858–868. doi:10.1111/j.0022-3646.1996.00858.x
Lewis J, Medlin LK, Raine R (2012) MIDTAL (Microarrays for the Detection of Toxic Algae): A protocol for a successful microarray hybridisation and analysis. Koeltz, Königstein, Germany
Loy A, Bodrossy L (2006) Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Act 363:106–219. doi:10.1016/j.cccn.2005.05.041
Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer K-H, Wagner M (2002) Oligonucleotide microarray for 16 S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. App Env Microbiol 68:5064–5081. doi:10.1128/AEM.68.10.5064-5081.2002
Loy A, Schulz C, Lücker S, Schöpfer-Wendels A, Stoecker K, Baranyi C, Lehner A, Wagner M (2005) 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order “Rhodocyclales”. Appl Environ Microbiol 71:1373–1386. doi:10.1128/AEM.71.3.1373-1386.2005
Marcelino LA, Backman V, Donaldson A, Steadman C, Thompson JR, Preheim SP, Lien C, Lim E, Veneziano D, Polz MF (2006) Accurately quantifying low-abundant targets amid similar sequences by revealing hidden correlations in oligonucleotide microarray data. PNAS 103:13629–13634. doi:10.1073/pnas.0601476103
Massana R, Pernice M, Bunge JA, del Campo J (2011) Sequence diversity and novelty of natural assemblages of picoeukaryotes from the Indian Ocean. ISME J 5:184–195. doi:10.1038/ismej.2010.104
Metfies K, Medlin LK (2005) Ribosomal RNA probes and microarrays: their potential use in assessing microbial biodiversity. Meth Enzymol 395:258–278. doi:10.1111/j.0022-3646.1996.00858
Metfies K, Borsutzki P, Gescher C, Medlin LK, Frickenhaus S (2008) PhylochipAnalyser—a program for analysing hierarchical probe sets. Mol Ecol Resour 8:99–102. doi:10.1111/j.1471-8286.2007.01927.x
Not F, Simon N, Biegala IC, Vaulot D (2002) Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat Microb Ecol 28:157–166. doi:10.3354/ame028157
Palmer C, Bik EM, Eisen MB, Eckburg PB, Sana TR, Wolber PK, Relman DA, Brown PO (2006) Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res 34:e5. doi:10.1093/nar/gnj007
Penna A, Bertozzini E, Battocchi C, Galluzzi L, Giacobbe MG, Vila M, Garces E, Luglie A, Magnani M (2006) Monitoring of HAB species in the Mediterranean Sea through molecular methods. J Plankton Res 29:19–38. doi:10.1093/plankt/fbl053
Peplies J, Lau SCK, Pernthaler J, Amann R, Glöckner FO (2004) Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ Microbiol 6:638–645. doi:10.1111/j.1462-2920.2004.00588.x
Scholin CA, Buck KR, Britschgi T, Cangelosi G, Chavez FP (1996) Identification of Pseudo-nitzschia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia 35:190–197. doi:10.2216/i0031-8884-35-3-190.1
Simon N, Campbell L, Ornolfsdottir E, Groben R, Guillou L, Lange M, Medlin LK (2000) Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J Euk Microbiol 47:76–84. doi:10.1111/j.1550-7408.2000.tb00014.x
Small J, Call DR, Brockman FJ, Straub TM, Chandler DP (2001) Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl Environ Microbiol 67:4708–4716. doi:10.1128/AEM.67.10.4708-4716.2001
Wagner M, Smidt H, Loy A, Zhou J (2007) Unravelling microbial communities with DNA-microarrays: challenges and future directions. Microb Ecol 53:498–506. doi:10.1007/s00248-006-9197-7
Acknowledgments
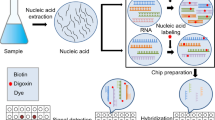
The members of the MIDTAL consortium (http://www.midtal.com/partners.php) are acknowledged for helpful discussions and support, for providing calibration data, for testing the software, and for critical comments on the manuscript especially Jixin Chen, Linda K Medlin, Johannes Hagstrøm, Jessica Kegel, Joe Taylor, Gary McCoy, and Lucia Barra. Linda K Medlin and Jessica Kegel provided the hierarchy file used as a basis for the file included in the GPR-Analyzer package. Some of the artwork used in Fig. 1 was provided courtesy of the Integration and Application Network, University of Maryland Center for Environmental Science (http://ian.umces.edu/symbols). This work was funded by the EU’s 7th Framework Program MIDTAL (FP7-ENV-2007-1-MIDTAL-201724).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Dittami, S.M., Edvardsen, B. GPR-Analyzer: a simple tool for quantitative analysis of hierarchical multispecies microarrays. Environ Sci Pollut Res 20, 6808–6815 (2013). https://doi.org/10.1007/s11356-012-1051-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-012-1051-5