Abstract
Introduction
Although sarcopenia greatly affects health and quality of life in older people, its pathophysiological causes are not fully elucidated. To face this challenge, omics technologies can be used. The metabolome gives a vision of the interaction between the genome and the environment through metabolic networks, thus contributing in clarifying the pathophysiology of the sarcopenic phenotype.
Objectives
The main goal of this study was to compare the plasma metabolome of sarcopenic and non-sarcopenic older people.
Methods
Cross-sectional study of 20 sarcopenic and 21 non-sarcopenic older subjects with available frozen plasma samples. Non-targeted metabolomic study by ultra-high-performance liquid chromatography–electrospray ionization tandem mass spectrometry (UHPLC-ESI–MS/MS) analysis with later bioinformatics data analysis. Once the significantly different metabolites were identified, the KEGG database was used on them to establish which were the metabolic pathways mainly involved.
Results
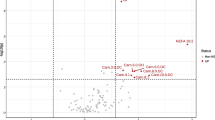
From 657 features identified, 210 showed significant differences between the study groups, and 30 had a FoldChangeLog2 > 2. The most interesting metabolic pathways found with the KEGG database were the biosynthesis of amino acids, arginine and proline metabolism, the biosynthesis of alkaloids derived from ornithine, linoleic acid metabolism, and the biosynthesis of unsaturated fatty acids.
Conclusions
The study results allowed us to confirm that the concept of “sarcopenic phenotype” is also witnessed at the plasma metabolite levels. The non-targeted metabolomics study can open a wide view of the sarcopenic features changes at the plasma level, which would be linked to the sarcopenic physiopathological alterations.


Similar content being viewed by others
Data availability
The study raw data was deposited in MetaboLights repository ULR: www.ebi.ac.uk/metabolights/MTBLS2679
References
Abbatecola, A. M., et al. (2011). Discovering pathways of sarcopenia in older adults: A role for insulin resistance on mitochondria dysfunction. The Journal of Nutrition, Health & Aging, 15, 890–895. https://doi.org/10.1007/s12603-011-0366-0
Adachi, Y., et al. (2018). Plasma amino acid profile in severely frail elderly patients in Japan. International Journal of Gerontology, 12, 290–293. https://doi.org/10.1016/j.ijge.2018.03.003
Albala, C., Sánchez, H., Lera, L., Angel, B., & Cea, X. (2011). Efecto sobre la salud de las desigualdades socioeconómicas en el adulto mayor: Resultados basales del estudio expectativa de vida saludable y discapacidad relacionada con la obesidad (Alexandros). Revista Médica De Chile, 139, 1276–1285. https://doi.org/10.4067/S0034-98872011001000005
Argilés, J. M., Campos, N., Lopez-Pedrosa, J. M., Rueda, R., & Rodriguez-Mañas, L. (2016). Skeletal muscle regulates metabolism via interorgan crosstalk: Roles in health and disease. Journal of the American Medical Directors Association, 17, 789–796. https://doi.org/10.1016/j.jamda.2016.04.019
Beermann, C., Jelinek, J., Reinecker, T., Hauenschild, A., Boehm, G., & Klör, H. U. (2003). Short term effects of dietary medium-chain fatty acids and n-3 long-chain polyunsaturated fatty acids on the fat metabolism of healthy volunteers. Lipids in Health and Disease, 2, 10. https://doi.org/10.1186/1476-511X-2-10
Bender, D. A. (2012). Amino acids synthesized from aspartate: Lysine, methionine (and cysteine), threonine and isoleucine. In D. A. Bender (Ed.), Amino acid metabolism (pp. 225–277). Chichester: Wiley.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Bodine, S. C., et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science, 294, 1704. https://doi.org/10.1126/science.1065874
Breen, L., & Phillips, S. M. (2011). Skeletal muscle protein metabolism in the elderly: Interventions to counteract the ‘anabolic resistance’ of ageing. Nutrition & Metabolism, 8, 68. https://doi.org/10.1186/1743-7075-8-68
Cai, D., Lee, K. K. H., Li, M., Tang, M. K., & Chan, K. M. (2004). Ubiquitin expression is up-regulated in human and rat skeletal muscles during aging. Archives of Biochemistry and Biophysics, 425, 42–50. https://doi.org/10.1016/j.abb.2004.02.027
Chabowski, A., et al. (2005). The subcellular compartmentation of fatty acid transporters is regulated differently by insulin and by AICAR. FEBS Letters, 579, 2428–2432. https://doi.org/10.1016/j.febslet.2004.11.118
Chaleckis, R., Naz, S., Meister, I., & Wheelock, C. E. (2018). LC-MS-based metabolomics of biofluids using all-ion fragmentation (AIF) acquisition. In M. Giera (Ed.), Clinical metabolomics: Methods and protocols (pp. 45–58). Springer New York.
Cole, J. T. (2015). Metabolism of BCAAs. In R. Rajendram, V. R. Preedy, & V. B. Patel (Eds.), Branched chain amino acids in clinical nutrition (Vol. 1, pp. 13–24). Springer, New York.
Cruzat, V., & Tirapegui, J. (2015). Glutamine and skeletal muscle. In R. Rajendram, V. R. Preedy, & V. Patel (Eds.), Glutamine in clinical nutrition (p. 12). Springer.
Cruz-Jentoft, A. J., et al. (2010). Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age and Ageing, 39, 412–423. https://doi.org/10.1093/ageing/afq034
Cruz-Jentoft, A. J., et al. (2019). Sarcopenia: Revised European consensus on definition and diagnosis. Age and Ageing, 48, 16–31. https://doi.org/10.1093/ageing/afy169
Curcio, F., et al. (2016). Biomarkers in sarcopenia: A multifactorial approach. Experimental Gerontology, 85, 1–8. https://doi.org/10.1016/j.exger.2016.09.007
Darmaun, D., Matthews, D. E., & Bier, D. M. (1988). Physiological hypercortisolemia increases proteolysis, glutamine, and alanine production. American Journal of Physiology-Endocrinology and Metabolism, 255, E366–E373. https://doi.org/10.1152/ajpendo.1988.255.3.E366
Das, U. N. (2018). Ageing: Is there a role for arachidonic acid and other bioactive lipids? A review. Journal of Advanced Research, 11, 67–79. https://doi.org/10.1016/j.jare.2018.02.004
Fan, J., Kou, X., Jia, S., Yang, X., Yang, Y., & Chen, N. (2016). Autophagy as a potential target for sarcopenia. Journal of Cellular Physiology, 231, 1450–1459. https://doi.org/10.1002/jcp.25260
Fischer, C., & Meese, C. O. (1985). Simultaneous determination of 6-oxo-prosta-glandin F1α and 2,3-dinor-6-oxo-prostaglandin F1α in biological fluids by stable isotope dilution and negative ion chemical ionization mass spectrometry. Biomedical Mass Spectrometry, 12, 399–404. https://doi.org/10.1002/bms.1200120808
Frontera, W. R., & Ochala, J. (2015). Skeletal muscle: A brief review of structure and function. Calcified Tissue International, 96, 183–195. https://doi.org/10.1007/s00223-014-9915-y
Fujita, S., Glynn, E. L., Timmerman, K. L., Rasmussen, B. B., & Volpi, E. (2009). Supraphysiological hyperinsulinaemia is necessary to stimulate skeletal muscle protein anabolism in older adults: Evidence of a true age-related insulin resistance of muscle protein metabolism. Diabetologia, 52, 1889–1898. https://doi.org/10.1007/s00125-009-1430-8
Gil, A., et al. (2018). One- vs two-phase extraction: Re-evaluation of sample preparation procedures for untargeted lipidomics in plasma samples. Analytical and Bioanalytical Chemistry, 410, 5859–5870. https://doi.org/10.1007/s00216-018-1200-x
Gomes, M. D., Lecker, S. H., Jagoe, R. T., Navon, A., & Goldberg, A. L. (2001). Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proceedings of the National Academy of Sciences of the United States of America, 98, 14440–14445. https://doi.org/10.1073/pnas.251541198
Hackshaw, A. (2008). Small studies: Strengths and limitations. European Respiratory Journal, 32, 1141. https://doi.org/10.1183/09031936.00136408
Harris, W. S., & Jacobson, T. A. (2009). CHAPTER 27: Omega-3 fatty acids. In C. M. Ballantyne (Ed.), Clinical lipidology (pp. 326–338). W.B. Saunders.
Hasten, D. L., Pak-Loduca, J., Obert, K. A., & Yarasheski, K. E. (2000). Resistance exercise acutely increases MHC and mixed muscle protein synthesis rates in 78–84 and 23–32 yr olds. American Journal of Physiology-Endocrinology and Metabolism, 278, E620–E626. https://doi.org/10.1152/ajpendo.2000.278.4.E620
Havel, R. J. (1987). Lipid transport function of lipoproteins in blood plasma. American Journal of Physiology-Endocrinology and Metabolism, 253, E1–E5. https://doi.org/10.1152/ajpendo.1987.253.1.E1
He, Y., et al. (2010). Glutamine synthetase in muscle is required for glutamine production during fasting and extrahepatic ammonia detoxification. The Journal of Biological Chemistry, 285, 9516–9524. https://doi.org/10.1074/jbc.M109.092429
Howard, B. V. (1999). Insulin resistance and lipid metabolism. The American Journal of Cardiology, 84, 28–32. https://doi.org/10.1016/S0002-9149(99)00355-0
Ivanisevic, J., & Thomas, A. (2018). Metabolomics as a tool to understand pathophysiological processes. In M. Giera (Ed.), Clinical metabolomics: Methods and protocols (pp. 3–28). Springer, New York.
Josse, J., & Husson, F. (2012). Selecting the number of components in principal component analysis using cross-validation approximations. Computational Statistics & Data Analysis, 56, 1869–1879. https://doi.org/10.1016/j.csda.2011.11.012
Karp, P. D., et al. (2019). The BioCyc collection of microbial genomes and metabolic pathways. Briefings in Bioinformatics, 20, 1085–1093. https://doi.org/10.1093/bib/bbx085
Katoh, A., et al. (2020). Long-chain fatty acids in sarcopenia patients with cardiovascular diseases: Importance of n-9 monounsaturated fatty acids. JCSM Clinical Reports. https://doi.org/10.1002/crt2.27
Kelton, J. G., & Blajchman, M. A. (1980). Prostaglandin I2 (prostacyclin). Canadian Medical Association Journal, 122, 175–179.
Kim, I.-Y., Park, S., Jang, J., & Wolfe, R. R. (2020). Understanding muscle protein dynamics: Technical considerations for advancing sarcopenia research. Journal of Korean Geriatric Society. https://doi.org/10.4235/agmr.20.0041
Kramer, I. F., et al. (2017). Both basal and post-prandial muscle protein synthesis rates, following the ingestion of a leucine-enriched whey protein supplement, are not impaired in sarcopenic older males. Clinical Nutrition, 36, 1440–1449. https://doi.org/10.1016/j.clnu.2016.09.023
Lê, S., Josse, J., & Husson, F. (2008). FactoMineR: An R package for multivariate analysis. Journal of Statistical Software, 1(1), 2008. https://doi.org/10.18637/jss.v025.i01
Lelo, A., Miners, J. O., Robson, R., & Birkett, D. J. (1986). Assessment of caffeine exposure: Caffeine content of beverages, caffeine intake, and plasma concentrations of methylxanthines. Clinical Pharmacology & Therapeutics, 39, 54–59. https://doi.org/10.1038/clpt.1986.10
Lera, L., et al. (2015). Estimación y validación de puntos de corte de índice de masa muscular esquelética para la identificación de sarcopenia en adultos mayores chilenos. Nutrición Hospitalaria, 31, 1187–1197. https://doi.org/10.3305/nh.2015.31.3.8054
Lera, L., et al. (2016). Prevalence of sarcopenia in community-dwelling Chilean elders according to an adapted version of the European Working Group on Sarcopenia in Older People (EWGSOP) criteria. The Journal of Frailty & Aging. https://doi.org/10.14283/jfa.2016.117
Lera, L., et al. (2018). Reference values of hand-grip dynamometry and the relationship between low strength and mortality in older Chileans. Clinical Interventions in Aging, 13, 317–324. https://doi.org/10.2147/CIA.S152946
Lu, D., et al. (2019). A systemic workflow for profiling metabolome and lipidome in tissue. Journal of Chromatography A, 1589, 105–115. https://doi.org/10.1016/j.chroma.2018.12.061
Lu, Y., et al. (2020). Systemic and metabolic signature of sarcopenia in community-dwelling older adults. The Journals of Gerontology: Series A, 75, 309–317. https://doi.org/10.1093/gerona/glz001
Masnoon, N., Shakib, S., Kalisch-Ellett, L., & Caughey, G. E. (2017). What is polypharmacy? A systematic review of definitions. BMC Geriatrics, 17, 230. https://doi.org/10.1186/s12877-017-0621-2
Mitchell, W., Atherton, P., Williams, J., Larvin, M., Lund, J., & Narici, M. (2012). Sarcopenia, dynapenia, and the impact of advancing age on human skeletal muscle size and strength: A quantitative review. Frontiers in Physiology, 3, 260. https://doi.org/10.3389/fphys.2012.00260
Moaddel, R., et al. (2016). Plasma biomarkers of poor muscle quality in older men and women from the Baltimore longitudinal study of aging. The Journals of Gerontology: Series A, 71, 1266–1272. https://doi.org/10.1093/gerona/glw046
Monnerat, G., et al. (2019). Metabolomic profiling suggests systemic signatures of premature aging induced by Hutchinson–Gilford progeria syndrome. Metabolomics, 15, 100. https://doi.org/10.1007/s11306-019-1558-6
Monnerat, G., et al. (2020). Different signatures of high cardiorespiratory capacity revealed with metabolomic profiling in elite athletes. International Journal of Sports Physiology and Performance, 15, 1156–1167. https://doi.org/10.1123/ijspp.2019-0267
Narici, M. V., & Maffulli, N. (2010). Sarcopenia: Characteristics, mechanisms and functional significance. British Medical Bulletin, 95, 139–159. https://doi.org/10.1093/bmb/ldq008
Nascimento, C. M., Ingles, M., Salvador-Pascual, A., Cominetti, M. R., Gomez-Cabrera, M. C., & Viña, J. (2019). Sarcopenia, frailty and their prevention by exercise. Free Radical Biology and Medicine, 132, 42–49. https://doi.org/10.1016/j.freeradbiomed.2018.08.035
Neu, J., Shenoy, V., & Chakrabarti, R. (1996). Glutamine nutrition and metabolism: Where do we go from here? The FASEB Journal, 10, 829–837. https://doi.org/10.1096/fasebj.10.8.8666159
Newsholme, E. A., & Parry-Billings, M. (1990). Properties of glutamine release from muscle and its importance for the immune system. Journal of Parenteral and Enteral Nutrition, 14, 63S-67S. https://doi.org/10.1177/014860719001400406
Ottestad, I., et al. (2018). Reduced plasma concentration of branched-chain amino acids in sarcopenic older subjects: A cross-sectional study. British Journal of Nutrition, 120, 445–453. https://doi.org/10.1017/S0007114518001307
Pence, H. E., & Williams, A. (2010). ChemSpider: An online chemical information resource. Journal of Chemical Education, 87, 1123–1124. https://doi.org/10.1021/ed100697w
Powell, W. S., & Rokach, J. (2015). Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochimica et Biophysica Acta, 1851, 340–355. https://doi.org/10.1016/j.bbalip.2014.10.008
Rennie, M. J., Tadros, L., Khogali, S., Ahmed, A., & Taylor, P. M. (1994). Glutamine transport and its metabolic effects. The Journal of Nutrition, 124, 1503S-1508S. https://doi.org/10.1093/jn/124.suppl_8.1503S
Sahebkar, A., et al. (2020). Pathophysiological mechanisms of statin-associated myopathies: Possible role of the ubiquitin-proteasome system. Journal of Cachexia, Sarcopenia and Muscle. https://doi.org/10.1002/jcsm.12579
Salek, R. M., Steinbeck, C., Viant, M. R., Goodacre, R., & Dunn, W. B. (2013). The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience, 2, 13–13. https://doi.org/10.1186/2047-217X-2-13
Santoru, M. L., et al. (2017). Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Scientific Reports, 7, 9523. https://doi.org/10.1038/s41598-017-10034-5
Scrimgeour, C. M., & Harwood, J. L. (2007). Fatty acid and lipid structure. In F. Gunstone, J. Harwood, & A. Dijkstra (Eds.), The lipid handbook with CD-ROM (3rd ed., pp. 1–36). Taylor & Francis Group.
Sengupta, A., & Narad, P. (2018). Metabolomics. In P. Arivaradarajan & G. Misra (Eds.), Omics approaches, technologies and applications: Integrative approaches for understanding OMICS data (pp. 75–97). Springer Singapore.
Shou, J., Chen, P.-J., & Xiao, W.-H. (2020). Mechanism of increased risk of insulin resistance in aging skeletal muscle. Diabetology & Metabolic Syndrome, 12, 14. https://doi.org/10.1186/s13098-020-0523-x
Spindler, S. R., Dhahbi, J. M., & Mote, P. L. (2003). Protein turnover, energy metabolism, aging, and caloric restriction. In M. P. Mattson (Ed.), Advances in cell aging and gerontology (Vol. 14, pp. 69–86). Elsevier.
Stangl, M. K., et al. (2019). Sarcopenia: Endocrinological and neurological aspects. Experimental and Clinical Endocrinology & Diabetes, 6, 8–22. https://doi.org/10.1055/a-0672-1007
Hamilton, P. B., & With the assistance of Rose Ruth. (1945). Glutamine: A major constituent of free α-amino acids in animal tissues and blood plasma. Journal of Biological Chemistry, 158, 397–409.
Tenenbaum, D., Maintainer, B. (2020). KEGGREST: Client-side REST acces to KEGG. R package version 1.30.1. In Bioconductor. http://www.bioconductor.org/packages/release/bioc/html/KEGGREST.html
Tipton, K. D., Hamilton, D. L., & Gallagher, I. J. (2018). Assessing the role of muscle protein breakdown in response to nutrition and exercise in humans. Sports Medicine (Auckland, NZ), 48, 53–64. https://doi.org/10.1007/s40279-017-0845-5
Volpi, E., Mittendorfer, B., Rasmussen, B. B., & Wolfe, R. R. (2000). The response of muscle protein anabolism to combined hyperaminoacidemia and glucose-induced hyperinsulinemia is impaired in the elderly. The Journal of Clinical Endocrinology and Metabolism, 85, 4481–4490. https://doi.org/10.1210/jcem.85.12.7021
Volpi, E., Sheffield-Moore, M., Rasmussen, B. B., & Wolfe, R. R. (2001). Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. JAMA, 286, 1206–1212. https://doi.org/10.1001/jama.286.10.1206
Wishart, D. S., et al. (2007). HMDB: The human metabolome database. Nucleic Acids Research, 35, D521–D526. https://doi.org/10.1093/nar/gkl923
Wu, G. Y., & Thompson, J. R. (1989). Is methionine transaminated in skeletal muscle? The Biochemical Journal, 257, 281–284. https://doi.org/10.1042/bj2570281
Yamada, M., et al. (2018). Plasma amino acid concentrations are associated with muscle function in older Japanese women. The Journal of Nutrition, Health & Aging, 22, 819–823. https://doi.org/10.1007/s12603-018-1014-8
Yannai, S. (2013). Dictionary of food compounds with CD-ROM. CRC Press.
Yaribeygi, H., Farrokhi, F. R., Butler, A. E., & Sahebkar, A. (2019). Insulin resistance: Review of the underlying molecular mechanisms. Journal of Cellular Physiology, 234, 8152–8161. https://doi.org/10.1002/jcp.27603
Zamboni, M., Gattazzo, S., & Rossi, A. P. (2019). Myosteatosis: A relevant, yet poorly explored element of sarcopenia. European Geriatric Medicine, 10, 5–6. https://doi.org/10.1007/s41999-018-0134-3
Zhao, Q., et al. (2018). A joint analysis of metabolomic profiles associated with muscle mass and strength in Caucasian women. Aging, 10, 2624–2635. https://doi.org/10.18632/aging.101574
Acknowledgements
We thank Dr. Fabricio Alves Barbosa da Silva, researcher of scientific computing program, Fundação Oswaldo Cruz, for his assistance in metabolomic pathways bioinformatic analysis, “network vision”. We acknowledge the Chilean National Fund for Scientific and Technological Development for the partial funding of this research (Fondecyt Grant 1130947 and Fondecyt Iniciación Grant 11190532).
Author information
Authors and Affiliations
Contributions
CA and RO conceptualized the study; GM, GRC, BA, CM, RO performed the experiments at different steps; LL, RO and GM performed the data analysis. CA, GM, BA, GRC, RO wrote the original draft. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Rafael Opazo, Bárbara Angel, Carlos Márquez, Lydia Lera, Gustavo R. Cardoso Dos Santos, Gustavo Monnerat, and Cecilia Albala declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11306_2021_1832_MOESM1_ESM.tiff
Supplementary file1—Fig. SF1 Score plot of principal component analysis (PCA) performed by Compound Discoverer 2.1 to evaluated the sample replicates reproducibility in the UHPLC-ESI-MS/MS analysis. PCA Dimension 1 versus Dimension 2 scores, with a 16.5% and 9.7% of the explained variances respectively. The blue spots are the total number of injections analyzed for the Sarcopenic group (three replicates of each sample); the orange spots are the total number of injections analyzed for the non-sarcopenic group (three replicates of each sample); the magenta spots are the QC-Pool samples in three replicates, and the green spots are the blank samples in three replicates. As it is possible to observe, the colored points are clearly grouped in differentiated clouds associated with the replicas of each experimental group, which allows us to propose that the analysis presented adequate reproducibility (TIFF 36719 kb)
11306_2021_1832_MOESM2_ESM.tiff
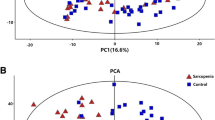
Supplementary file2—Fig. SF2 Score plot of principal component analysis (PCA) using 657 features identified by the metabolomic study between the study groups. PCA Dimension 1 versus Dimension 2 scores. The red points correspond to sarcopenic subjects, and the black points correspond to non-sarcopenic subjects (TIFF 1648 kb)
11306_2021_1832_MOESM3_ESM.tiff
Supplementary file3—Fig. SF3 Proportion bar chart of the sources of origin class for the 30 most differentiated plasma metabolites between the study groups. Only the metabolites with FoldchageLog2 equal to or greater than 2 (n = 28) were included in the analysis. The classification groups in descending order were endogenous/food, strictly food, drugs, herbicides, and non-determined (TIFF 1128 kb)
11306_2021_1832_MOESM4_ESM.tiff
Supplementary file4—Fig. SF4 Box and whisker plots of the unsaturated fatty acids with significant (padj ≤ 0.12) plasma level differences between non-sarcopenic and sarcopenic subjects. A) Omega 6 series, B) Omega 3 series, and C) Omega 9 and 7 series. The box shows the interquartile range (25th to 75th percentile), the blackline shows the median (50th percentile), and the whiskers show the maximum and minimum (TIFF 1758 kb)
11306_2021_1832_MOESM5_ESM.tiff
Supplementary file5—Fig. SF5 Box and whisker plot of the saturated fatty acids with significant (padj ≤ 0.12) plasma level differences between non-sarcopenic and sarcopenic subjects. The box shows the interquartile range (25th to 75th percentile), the blackline shows the median (50th percentile), and the whiskers show the maximum and minimum (TIFF 1289 kb)
11306_2021_1832_MOESM6_ESM.tiff
Supplementary file6—Fig. SF6 Box and whisker plots of A) glycolipid with significant (padj ≤ 0.12) plasma level differences between non-sarcopenic and sarcopenic subjects, and B) and phospholipid with significant (padj ≤ 0.12) plasma level differences between non-sarcopenic and sarcopenic subjects. The box shows the interquartile range (25th to 75th percentile), the blackline shows the median (50th percentile), and the whiskers show the maximum and minimum (TIFF 1758 kb)
Rights and permissions
About this article
Cite this article
Opazo, R., Angel, B., Márquez, C. et al. Sarcopenic metabolomic profile reflected a sarcopenic phenotype associated with amino acid and essential fatty acid changes. Metabolomics 17, 83 (2021). https://doi.org/10.1007/s11306-021-01832-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-021-01832-0