Abstract
Hedgerows are an important component of agricultural landscapes, but in recent years have increasingly faced threats such as habitat loss, land use change, climate change, invasive species, pests and plant pathogens. Given the potential importance of genetic diversity in countering these threats, and the spatial distribution of such diversity within and across natural populations, we analyzed levels and patterns of diversity in blackthorn (Prunus spinosa), a key component of many hedgerows. Twenty-one populations of blackthorn from a mixture of hedgerows and woodlands were genotyped for four nuclear and five chloroplast microsatellites. Three hundred twenty-one unique clonal genotypes were identified from 558 individuals analyzed, 207 of which were found in a single individual. With the exception of a single population that appears to have been planted recently from seed (Peatlands Park), all populations exhibited evidence of vegetative reproduction via suckering. Multi-ramet clones were highly spatially structured within populations, and ranged in size from < 1 to 258 m. These findings indicate that asexual reproduction is widespread in the populations of blackthorn studied. Although levels of clonality varied across study sites, there was clear spatial structuring of clones in each case. Such clonal organization should be taken into account in hedge management or where planting or replanting of hedgerows becomes necessary. Knowledge of the patterns and extent of spatial structuring of genotypes within potential source populations will allow the selection of genetically divergent material, rather than selection of clonal replicates of the same genotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hedgerows have been an integral part of agricultural landscapes for hundreds of years. They were planted initially to act as field boundaries, but are now considered important not just for their function as barriers, but also due to the diverse ecosystem services they provide (Montgomery et al. 2020). These include acting as windbreaks, preventing soil erosion, and acting as habitats, food sources and wildlife corridors for animals (Burel and Baudry 1995; Staley et al. 2012). It has also been suggested that hedgerows could play a key role in carbon sequestration, as well as reducing N2O emissions (Falloon et al. 2004). However, hedgerow lengths and volumes have declined in modern times: the 2007 UK Countryside Survey estimated a decrease of 6% in the length of managed hedgerows over the preceding decade (Carey et al. 2008). Further, as a result of management based on prescriptions related to agricultural subsidies, many hedges are reduced in size and cut on an annual basis (Staley et al. 2015). There is also a risk that these declines may be compounded by the ongoing threats faced by both woodland and hedgerow species such as habitat loss, land use change, climate change, invasive species, pests and plant pathogens (Rackham 2008; Spaans et al. 2018).
Prunus spinosa (blackthorn) is a small, deciduous shrub that is a key component of many hedgerows, as well as being found in forests throughout Europe. In Britain, it is estimated to be present in around half of all hedges, where it acts as both habitat and as a food source for a wide range of animals (Wilson 2019). Its thorny branches protect nesting animals such as birds, whilst the hermaphrodite flowers provide nectar for a range of pollinator species, and berries (sloes) are eaten by birds and mammals, thus facilitating seed dispersal (Yeboah and Woodell 1987; Guitian et al. 1993). The species has a wide distribution across Europe, and extends into the southern part of the Scandinavian Peninsula and the northern part of the Iberian Peninsula (Popescu and Caudullo 2016). It is an allotetraploid (Reynders-Aloisi and Grellet 1994), and is generally thought to be self-incompatible, with a self-incompatibility locus under genetic control often found in the genus Prunus, including P. spinosa (Yeboah and Woodell 1987; Nunes et al. 2006; Halász et al. 2017). Self-pollination has been reported, however, but with fewer fruits produced than in crosses (Guitian et al. 1993). Natural vegetative reproduction occurs extensively in blackthorn via suckering, with evidence that blackthorn clones can grow to a few tens of metres (Leinemann et al. 2010, 2014). If clonal reproduction plays a major role in the establishment and spread of P. spinosa hedgerows and/or woodlands, this could limit overall genetic diversity, which in turn limits the adaptive potential of the species to react to potential threats highlighted above (Frankham 2005). Furthermore, should the need arise to replant or supplement hedgerows, knowledge of the patterns and extent of spatial structuring of genotypes within potential source populations will allow the selection of genetically divergent material, rather than selection of clonal replicates of the same genotype.
The aim of the present study was to examine the extent and structure of vegetative reproduction in blackthorn hedgerows and woodlands, since a previous study on the other key hedgerow species, hawthorn, from the same region (Ireland) surprisingly showed no evidence of clonality (Brown et al. 2016a). We utilized high-resolution nuclear and chloroplast microsatellites in combination with GPS to identify clonal individuals to determine levels of clonality and overall genetic diversity, and to map patterns of vegetative spread.
Materials and methods
Sampling and DNA extraction
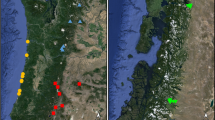
Samples were collected from 18 sites in Northern Ireland along with three sites in the Republic of Ireland (Fig. 1). Samples were taken from a combination of hedgerows and woodland. Sites were selected that had been designated previously as ancient or semi-natural based on data collected for the Woodland Trust Inventory of ancient and long-established woodland in Northern Ireland (www.backonthemap.org.uk) and the National Survey of Native Woodlands 2003–08 in the Republic of Ireland (www.npws.ie). Government information from the Department of the Environment such as ASSIs (Areas of Special Scientific Interest), as well as the landscape character areas listing the woodlands and species present in each region (https://www.doeni.gov.uk), was also used to identify potential sites. Some sample sites were discovered opportunistically, being clearly visible from roads. For sampling, a number of leaves were taken from each of a minimum of 30 individual trees representing the distribution of P. spinosa across the study site and stored in silica gel. The GPS coordinates of each individual sampled were recorded at the same time. DNA was extracted from a single leaf punched disc using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987).
Locations of sites sampled in this study. Numbers correspond to those in Table 1
Genotyping
Samples were genotyped for four nuclear microsatellite loci and five chloroplast microsatellite loci. Three of the four nuclear loci (BPPCT007, BPPCT025 and BPPCT037) were originally described in Dirlewanger et al. (2002), whilst the other (CPSCT021) was originally described in Mnejja et al. (2004). All four had been demonstrated previously to be moderately to highly polymorphic in P. spinosa (Halász et al. 2017). Forward primers included a 19 bp M13 tail (CACGACGTTGTAAAACGAC) and reverse primers included a 7 bp tail (GTGTCTT). PCR was carried out in a total volume of 10 μl containing 100 ng genomic DNA, 5 pmol of 6-FAM labelled M13 primer, 0.05 pmol of each M13-tailed forward primer, 5 pmol each reverse primer, 1 × PCR reaction buffer, 200 μM each dNTP, 2.5 mM MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega, Sunnyvale, CA, USA). PCR was carried out using the following conditions: initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C (BPPCT007, BPPCT025, BPPCT037) or 46 °C (CPSCT021) for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. Genotyping was carried out on an AB3730xl capillary genotyping system (Applied Biosystems, Foster City, CA, USA). Allele sizes were scored using the GeneMarker software (V1.8, SoftGenetics).
Chloroplast microsatellites were developed by searching P. spinosa GenBank sequences for mononucleotide repeats (Provan et al. 2001). Primers were designed using the Primer3 program (Untergasser et al. 2012) to amplify five loci in two multiplex reactions (AM282681/HQ244364 and KJ994356.1/KJ994356.2/AY500658; Table S1). PCR and genotyping were carried out as described above, except that 30 cycles were performed with an annealing temperature of 58 °C.
Data analysis
Individuals (ramets) sharing multi-locus genotypes (MLGs) were considered to be clones (genets). Because P. spinosa is polyploid, it was not possible to score genotypes based on allele frequencies, and, consequently, we could not use the standard method of Parks and Werth (1993) to calculate the probabilities of each MLG arising through sexual reproduction. Nevertheless, given that MLGs were characterized by extremely high levels of heterozygosity at three highly variable loci (BPPCT007 [33 alleles], BPPCT025 [36 alleles] and CPSCT021 [37 alleles]), it is extremely unlikely that segregation and recombination associated with sexual reproduction (either biparentally or through selfing) would give rise to identical heterozygous MLGs (Tibayrenc et al. 1990). Similarly, in the absence of allele frequencies, we were unable to carry out several standard population genetic analyses (e.g. HE, AMOVA) based on the nuclear data. Levels of genetic diversity (R) were measured as genotypic richness (R = [G-1]/[N-1]), where G was the number of genets identified, and N was the number of ramets sampled, clonal diversity (D*), which is the equivalent of Simpson’s Diversity Index (Simpson 1949), and observed heterozygosity (HO), which was calculated as the proportion of heterozygous genets averaged over loci. For analysis of chloroplast data, a single representative of each nuclear MLG was used. Levels of chloroplast variation were estimated as gene diversity (H), calculated using the Arlequin software package (V3.5.1.2; Excoffier and Lischer 2010). The level of population differentiation for nuclear markers (FST) was calculated using the polysat software package (Clark and Jasieniuk 2011), and differentiation based on chloroplast haplotypes (ΦST) calculated using Arlequin. To test for isolation-by-distance (FST / [1 – FST] vs. geographical distance), a Mantel test was carried out using the ISoLDE package implemented in Genepop (V4.7.5; Rousset 2008). Again, a single representative of each nuclear MLG was used, and population-pairwise FST values were calculated using polysat. Genet lengths were estimated in QGIS (V3.16.2).
Results
Nuclear genotypes were obtained for 588 individuals in total. Between nine (BPPCT037) and 37 (CPSCT021) alleles were observed at the four loci examined (mean = 28.75 per locus), giving 321 unique genets (Tables S2, S3). Of these, 207 were found in a single individual. No genets were shared across populations. Genets tended to be highly spatially structured within sites, with little overlap (for examples, see Fig. 2; all maps of genets within each location studied are presented in Figure S1), and ranged in size from < 1 to 258 m (mean = 50 m; median = 31 m; Fig. 3). Levels of genotypic richness (R), clonal diversity (D*) and observed heterozygosity (HO) ranged from 0.200 (Warrenpoint) to 1.000 (Peatlands Park; mean = 0.533), from 0.727 (Aghanloo Wood) to 1.000 (Peatlands Park; mean = 0.906) and from 0.720 (Sloughan Glen) to 0.964 (Glenarm; mean = 0.817), respectively (Table 1).
Between three (AY500685) and six (KJ994356.2), alleles were observed at the five chloroplast microsatellite loci studied (Table S1). Combining alleles gave rise to 27 different haplotypes (Tables S2 and S4). Levels of chloroplast haplotype diversity (H) ranged from zero (Woodburn, Aghanloo Wood and Strabane Glen) to 0.846 (Peatlands Park; mean = 0.454; Table 1). Excluding Aghanloo Wood and Banagher Glen, which contained samples from a mixture of hedgerows and woodland, and Peatlands Park, which most likely was planted from seed relatively recently (see Discussion), there was no significant difference between levels of genetic diversity between hedgerows and woodlands based on any of the measures (Fig. 4). Levels of population differentiation were estimated as FST = 0.043 for the nuclear microsatellites (FST = 0.042 excluding Peatlands Park), and ΦST = 0.254 for the chloroplast microsatellites (ΦST = 0.230 excluding Peatlands Park). Population-pairwise FST values ranged from 0.009 (Ness Wood vs. Sloughan Glen) to 0.061 (Glenarm vs. Sloughan Glen; Table S5). No evidence of isolation-by-distance was observed (Mantel Test P = 0.266; Figure S2).
Discussion
The findings of the present study indicate that asexual reproduction is widespread in the populations of blackthorn studied. Although the levels of clonality varied across the study sites, there was clear spatial structuring of clones within each location, with some clones extending over tens and even hundreds of metres. Such clonal organization and the extent of clonal spread should be taken into account if conservation, management or planting of hedgerows is considered necessary.
Of the few population genetic studies carried out previously on blackthorn, only two have studied levels of clonality. Leinemann et al. (2010) primarily used allozymes to examine natural populations from Germany and found that only 4–10% of individuals analyzed exhibited unique genotypes, compared with 35% in the present study. These low values may at least be in part due to the limited resolving power of allozymes relative to microsatellites, since the addition of two microsatellite loci to the allozyme data for one of the populations saw an increase in the proportion of unique MLGs from 10 to 29%, which is more in line with the levels observed here. Similarly, the values for R in the natural populations studied (0.12–0.56; mean = 0.31) were generally lower in the German study than those found in our study (Table 1). A more recent study using amplified fragment length polymorphism (AFLP) in populations from Germany, Italy and Hungary (Leinemann et al. 2014), however, found higher values for R (0.49–1.00; mean = 0.78), reflecting the multi-locus nature of the AFLP technique, as did a similar study in Belgium (Mijnsbrugge et al. 2013). These values of R are more in keeping with the current study (Table 1). The primary aim of these studies, however, was to identify genetically diverse plant material for collection, rather than the analysis of clonal individuals per se.
Estimated clone sizes (length) in the present study ranged from less than 1 to 258 m. The majority were of a similar size to the mean values of 12, 27 and 30 m reported by Leinemann et al. (2010) and the largest clones (30–40 m) observed by Leinemann et al. (2014), but several were much larger, with 14 clones over 100 m in length. Clones of more than a few metres in size are far more likely to have arisen through natural vegetative reproduction via suckering than through management practices such as laying or coppicing (Staley et al. 2015), which historically have not been carried out widely in Ireland (Pilcher & Hall 2001). The high degree of spatial structuring observed across all sites is consistent with the “phalanx” growth form, wherein there is no mixing of genets, and ramets are packed closely together due to connections between them having few or no internodes. This is contrast to a “guerrilla” form where clones are not only more spread out from one another due to longer connections, but where there is also mixing of clones (Lovett Doust 1981). It is also possible that those genotypes which are more successful can outcompete other less successful genotypes and replace or displace them over time, and that this, combined with limited sexual recruitment, can result in the formation of clonal stands (Eriksson 1993).
The population sampled from Peatlands Park, County Armagh, differed from all others in the study in that each individual sampled exhibited a unique genotype (Figure S1.6). The park is a visitor attraction, comprising a mixture of managed and semi-natural woodland and peatland habitats, and the samples were collected from a hedge that runs alongside a railway line used by a small train that takes visitors through the park. As such, it is likely that these plants were planted from seed relatively recently. This is supported by the fact that Peatlands Park was the only population that did not exhibit chloroplast haplotype 1, by far the most common across all populations studied. Furthermore, only a single individual exhibited a haplotype found elsewhere (Murlough Bay), the remaining individuals being represented by seven population-specific haplotypes. A similar, if less pronounced, scenario is seen in the Ness Wood population (Figure S1.13), where all the sampled individuals from the road leading to the “entrance” to the wood exhibited unique genets. Unlike in Peatlands Park, however, they all share the most common chloroplast haplotype, which suggests that if they have been planted from seeds, these most likely had “local” provenance.
The level of population genetic differentiation based on nuclear markers (FST = 0.043; 0.042 excluding Peatlands Park) was similar to that observed previously using allozymes (average Dj = 0.065; Kleinschmit et al. 2008), but lower than that found in the previous AFLP studies (0.21 in Mijnsbrugge et al. 2013 and 0.15 in Leinemann et al. 2014). This may reflect the fact that dominant markers such as AFLP tend to generate higher levels of population differentiation than codominant markers (Isabel et al. 1999). The value for blackthorn in the present study was generally higher than levels observed in other native tree species that have been studied in Ireland using microsatellites, including hawthorn (0.009; Brown et al. 2016a), ash (0.013; Beatty et al. 2015a), hazel (0.017; Brown et al. 2016b) and alder (0.020; Beatty et al. 2015b), but lower than that in juniper (0.096; Provan et al. 2008). This is probably due at least in part to the clonal nature of most of the populations studied, with reduced cross-pollination, seed production and subsequent dispersal relative to the other species. Differentiation exhibited by chloroplast markers was higher (ΦST = 0.254; 0.230 excluding Peatlands Park), consistent with the maternal (i.e. via seed) transmission of the chloroplast genome. This value is similar to those found across a similar geographical area (0.30; Leinemann et al. 2014), as well as across Europe (0.195 in Mohanty et al. 2000 and 0.30 in Mohanty et al. 2002). It is also comparable to levels found across Ireland for other tree species such as alder (0.186; Beatty et al. 2015b), juniper (0.249; Provan et al. 2008) and ash (0.263; Beatty et al 2015a), but higher than in hawthorn (0.039; Brown et al. 2016a). In all cases, populations were generally characterized by one or two dominant chloroplast haplotypes, reflecting the founder effects associated with postglacial recolonization (Provan and Bennett 2008).
The significant role of vegetative reproduction in the population genetic structure of blackthorn has several implications for management and conservation. Although hedges and woodlands, in most cases, comprise multiple clones, the spatial organization of clonal units should be considered when selecting material for transplantation. To maximize genetic variation when sampling, individuals ideally should be collected several tens of metres apart, particularly within hedges, to minimize the chance of repeated sampling of the same genet. The current research suggests that these hedges may represent an important repository for much more ancient genetic material which survived the clearance of primary and secondary forest from Neolithic times onwards. In addition, the comparable levels of genetic diversity in both hedgerows and woodlands means that hedges, as well as more spatially extensive woodlands, might represent valuable reservoirs of diversity, in addition to their key provision of a range of ecosystem services. It is vital, therefore, that diversity of both populations and genetic diversity within these populations is maintained by sympathetic management of field boundaries.
Data availability statement
Data will be deposited in Dryad on acceptance, and are available from the Corresponding Author on request.
Change history
22 July 2022
Handling editor name correction.
References
Beatty GE, Brown JA, Cassidy EM et al (2015a) Lack of genetic structure and evidence for long-distance dispersal in ash (Fraxinus excelsior) populations under threat from an emergent fungal pathogen: implications for restorative planting. Tree Genet Genomes 11:53
Beatty GE, Montgomery WI, Tosh DG, Provan J (2015b) Genetic provenance and best practice woodland management: a case study in native alder (Alnus glutinosa). Tree Genet Genomes 11:92
Brown JA, Beatty GE, Montgomery WI, Provan J (2016b) Broad-scale genetic homogeneity in natural populations of common hazel (Corylus avellana) in Ireland. Tree Genet Genomes 12:122
Brown JA, Beatty GE, Finlay CMV, Montgomery WI, Tosh DG, Provan J (2016a) Genetic analyses reveal high levels of seed and pollen flow in hawthorn (Crataegus monogyna Jacq.), a key component of hedgerows. Tree Genet Genomes 12:58.
Burel F, Baudry J (1995) Social, aesthetic and ecological aspects of hedgerows in rural landscapes as a framework for greenways. Landscape Urban Plann 33:327–340
Carey PD, Wallis S, Chamberlain PM et al. (2008) Boundary and linear features. In: Carey PD, Wallis S, Chamberlain PM et al. (eds) Countryside Survey: UK Results from 2007. Centre for Ecology and Hydrology, pp50–60.
Clark LV, Jasieniuk M (2011) polysat: an R package for polyploid microsatellite analysis. Mol Ecol Resources 11:562–566
Dirlewanger E, Cosson P, Tavaud M et al (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull 19:11–15
Eriksson O (1993) Dynamics of genets in clonal plants. Trends Ecol Evol 8:313–316
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resources 10:564–567
Falloon P, Powlson D, Smith P (2004) Managing field margins for biodiversity and carbon sequestration: a Great Britain case study. Soil Use Managem 20:240–247
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Guitian J, Guitian P, Sanchez JM (1993) Reproductive biology of two Prunus species (Rosaceae) in the Northwest Iberian Peninsula. Plant Syst Evol 185:153–165
Halász J, Makovics-Zsohár N, Szőke F, Ercisli S, Hegedűs A (2017) Simple sequence repeat and S-locus genotyping to explore genetic variability in polyploid Prunus spinosa and P. insititia. Biochemical Genet 55:22–33
Isabel N, Beaulieu J, Thériault P, Bosquet J (1999) Direct evidence for biased gene diversity estimates from dominant random amplified polymorphic DNA (RAPD) fingerprints. Mol Ecol 8:477–483
Kleinschmit J, Leinemann L, Hosius B (2008) Gene conservation through seed orchards – a case study of Prunus spinosa L. In: Lindgren E (ed) Seed Orchards: Proceedings from a Conference at Umeå, Sweden, 26–28 September 2007. SLU/Publikationstjänst, Uppsala, Sweden, pp 115–125
Leinemann L, Steiner W, Hosius B, Kleinschmit J (2010) Klonale Reproduction in naturnahen Vorkommen der Schlehe (Prunus spinosa L.). Forstarchiv 81:165–169
Leinemann L, Kleinschmit J, Fussi B et al (2014) Genetic composition and differentiation of sloe (Prunus spinosa L.) populations in Germany with respect to the tracing of reproductive material. Plant Syst Evol 300:2115–2125
Lovett Doust L (1981) Population dynamics and local specialization in a clonal perennial (Ranunculus repens). I. The dynamics of ramets in contrasting habitats. J Ecol 69:743–755
Mijnsbrugge KV, Depypere L, Chaerle P, Goetghebeur P, Breyne P (2013) Genetic and morphological variability among autochthonous Prunus spinosa populations in Flanders (northern part of Belgium): implications for seed sourcing. Plant Ecol Evol 146:193–202
Mnejja M, Garcia J, Howad W, Badenes ML, Arús P (2004) Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol Ecol Notes 4:163–166
Mohanty A, Martin JP, Aguinagalde I (2000) Chloroplast DNA diversity within and among populations of the allotetraploid Prunus spinosa L. Theor Appl Genet 100:1304–1310
Mohanty A, Martin JP, Aguinagalde I (2002) Population genetic analysis of European Prunus spinosa (Rosaceae) using chloroplast DNA markers. Am J Bot 89:1223–1228
Montgomery WI, Caruso T, Reid N (2020) Hedgerows as ecosystems: service delivery, management, and restoration. Annu Rev Ecol Evol Syst 51:81–102
Nunes MDS, Santos RAM, Ferreira SM, Vieira J, Vieira CP (2006) Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytol 172:577–587
Parks JC, Werth CR (1993) A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae). Am J Bot 80:537–544
Pilcher JR, Hall VA (2001) Flora Hibernica: the wild flowers, trees and plants of Ireland. Collins Press, Cork, Ireland.
Popescu I, Caudullo G (2016) Prunus spinosa in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A (eds) European Atlas of Forest Tree Species. e018f4e. Publications Office of the European Union, Luxembourge.
Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23:564–571
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147
Provan J, Beatty GE, Hunter AM et al (2008) Restricted gene flow in fragmented populations of a wind-pollinated tree. Conserv Genet 9:1521–1532
Rackham O (2008) Ancient woodlands: Modern threats. New Phytol 180:571–586
Reynders-Aloisi S, Grellet F (1994) Characterization of the ribosomal DNA units in two related Prunus species (P. cerasifera and P. spinosa). Plant Cell Rep 13:641–646
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resources 8:103–106
Simpson HE (1949) Measurement of diversity. Nature 163:688
Spaans F, Caruso T, Montgomery WI (2018) The abundance and condition of hedgerow tree standards in Northern Ireland. Biol Environ: Proc R Irish Acad 118:129–145
Staley JT, Sparks TH, Croxton PJ et al (2012) Long-term effects of hedgerow management policies on resource provision for wildlife. Biological Conserv 145:24–29
Staley JT, Amy SR, Adams NP, Chapman RE, Peyton JM, Pywell RF (2015) Re-structuring hedges: rejuvenation management can improve the long term quality of hedgerow habitats for wildlife in the UK. Biological Conserv 186:187–196
Tibayrenc M, Kjellberg F, Ayala FJ (1990) A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma and their medial consequences. Proc Natl Acad Sci USA 87:2414–2418
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3 - new capabilities and interfaces. Nucleic Acids Res 40:e115
Wilson PJ (2019) Botanical diversity in the hedges and field margins of lowland Britain. In: Dover JW (ed) The Ecology of Hedgerows and Field Margins. Routledge, UK.
Yeboah KG, Woodell SRJ (1987) Flowering phenology, flower colour and mode of reproduction of Prunus spinosa L. (Blackthorn); Crataegus monogyna Jacq. (Hawthorn); Rosa canina L. (Dog Rose); and Rubus fruticosus L. (Bramble) in Oxfordshire. England Functional Ecol 1:261–268
Acknowledgements
We are grateful to landowners for allowing access to their property, and to Sally Montgomery, Liz Brown, Lexi Brown and Jennifer McDowell for assistance with sampling.
Funding
James Brown’s PhD studentship was funded by the Department of Agriculture, Environment and Rural Affairs, Northern Ireland.
Author information
Authors and Affiliations
Contributions
JP and WIM conceived the study. JP designed the experiments. JAB and WIM collected material. JAB and JP carried out the labwork and analyzed the data. JAB drafted the manuscript, which was revised by JP and WIM.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Additional information
Communicated by N. Tomaru.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brown, J.A., Montgomery, W.I. & Provan, J. Strong spatial structuring of clonal genetic diversity within blackthorn (Prunus spinosa) hedgerows and woodlands. Tree Genetics & Genomes 18, 5 (2022). https://doi.org/10.1007/s11295-022-01538-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01538-x