Abstract
Background
Despite its implications for population dynamics and evolution, the relationship between genetic and phenotypic variation in wild populations remains unclear. Here, we estimated variation and plasticity in life-history traits and fitness of the annual plant Arabidopsis thaliana in two common garden experiments that differed in environmental conditions. We used up to 306 maternal inbred lines from six Iberian populations characterized by low and high genotypic (based on whole-genome sequences) and ecological (vegetation type) diversity.
Results
Low and high genotypic and ecological diversity was found in edge and core Iberian environments, respectively. Given that selection is expected to be stronger in edge environments and that ecological diversity may enhance both phenotypic variation and plasticity, we expected genotypic diversity to be positively associated with phenotypic variation and plasticity. However, maternal lines, irrespective of the genotypic and ecological diversity of their population of origin, exhibited a substantial amount of phenotypic variation and plasticity for all traits. Furthermore, all populations harbored maternal lines with canalization (robustness) or sensitivity in response to harsher environmental conditions in one of the two experiments.
Conclusions
Overall, we conclude that the environmental attributes of each population probably determine their genotypic diversity, but all populations maintain substantial phenotypic variation and plasticity for all traits, which represents an asset to endure in changing environments.
Similar content being viewed by others
Background
Although there are few studies monitoring populations for both long-term demographic and genetic dynamics [1], genetic variation is intuitively considered as a valuable attribute for population viability, particularly when populations are challenged by environmental fluctuations [2]. This view results from the positive relationship between genetic variation and population size [3, 4], both used as proxies for population viability. In particular, it is assumed that a population harboring a large pool of genetic variants will have greater chances of buffering the effects of environmental shifts, whereas populations impoverished genetically may see such buffering ability diminished. As selection acts upon phenotypes, a large pool of genetic variants in a population ought to be translated into a broad spectrum of phenotypes to make the buffering effect of genetic variation effective in changing environments. However, empirical data on the actual relationship between genetic and phenotypic variation in populations reveals that such a relationship is rather weak or inexistent [5,6,7,8,9,10]. Some explanations include past bottlenecks shaping molecular and quantitative variation in a different manner or differential environmental effects on molecular and quantitative traits [6]. Hence, the relationship between genetic and phenotypic variation and its utility to biologists for assessing population viability probably need to be reappraised.
In this study, we evaluated the relationship between genotypic diversity, i.e. the number and frequency of genotypes in a population, and phenotypic variation in life-history traits in wild populations of the self-fertilizing annual plant Arabidopsis thaliana. To increase the odds of studying populations not affected by recent disturbances, we selected populations from Iberian environments that remained undisturbed by land-use changes for several decades [11] and that are known to exist at least since the early 2000s, when we sampled most of them for the first time. Furthermore, the limited dispersal of A. thaliana [11,12,13,14,15] ensures that the genetic composition of populations was chiefly driven by mutation-selection balance. Here, we quantified variation in life-history traits and fitness of A. thaliana maternal inbred lines (maternal lines hereafter), estimated in common garden experiments over two consecutive years that differed in weather conditions. Hence, we also assessed phenotypic plasticity as another component of phenotypic performance displayed by populations [16,17,18], which may have important implications for population viability and evolutionary dynamics.
We tested two hypotheses to examine the relationship between genotypic diversity, estimated with whole-genome sequences, and phenotypic variation, estimated in common garden experiments, in A. thaliana populations. We took advantage of the fact that our study populations split into two groups: one with low and the other with high genotypic diversity. As populations with low genotypic diversity were located in edge environments, mostly determined by low (e.g. seaside location) and high (e.g. mountain passes) elevations across the species’ Iberian distribution, we expected to detect lower phenotypic variation in these populations as selection is expected to be stronger in edge environments [19, 20]. In particular, A. thaliana avoids the hot and dry summers at low Iberian elevations, whereas at high altitudes, the species has to overcome harsh winters. This expectation is supported by previous findings on the effects of elevation gradients on Iberian A. thaliana, indicating that edge environments are good predictors of genetic attributes (e.g. lower genetic diversity at higher altitudes; [21]), architectural traits (e.g. larger plant size at bolting at higher altitudes; [22]), fitness-related traits (e.g. weaker seed dormancy and later flowering time at higher altitudes; [23]), and demographic features (e.g. dominance of spring-germinated plants at higher altitudes; [24]). In contrast, populations with higher genotypic diversity in core environments at intermediate elevations could show the opposite trend if selection acted in a more relaxed manner.
In addition, our study populations with low genotypic diversity in edge environments were also characterized by lower ecological diversity, determined by the diversity of major vegetation types, than populations with high genotypic diversity in core environments. Thus, we also hypothesized that differences in the ecological diversity of populations promoted lower and higher phenotypic variation in populations with low and high genotypic diversity, respectively. On top of the effects of fine-scale selection on fitness-related traits in heterogeneous environments [25,26,27,28,29], which promote within-population phenotypic variation, phenotypic plasticity has also been seen to be enhanced in environments with higher levels of ecological heterogeneity [30,31,32]. In addition, environmental maternal effects may also provide transgenerational adaptive plasticity in plants [33,34,35,36]. Overall, we predicted that core populations with higher genotypic and ecological diversity exhibited, not only higher phenotypic variation, but higher phenotypic plasticity than those in edge environments.
Here, we asked (i) what is the pattern of phenotypic variation and plasticity in life-history traits and fitness estimated in common garden experiments of A. thaliana populations differing in genotypic and ecological diversity? And (ii) what are the ecological and genetic drivers of phenotypic variation and plasticity in this set of populations representing core and edge Iberian environments? We discuss the results in the context of the intertwined relationship between genetic and phenotypic variation to broaden our comprehension of the evolutionary dynamics of natural A. thaliana populations and their viability in changing environments.
Methods
Source populations and sampling
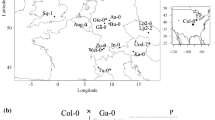
F.X. Picó, R. Gómez and C. Alonso-Blanco collected seeds from all individuals used in this study. It must be noted that A. thaliana is a common plant species not categorized as protected or endangered in any species list of the Convention on the Trade in Endangered Species of Wild Fauna and Flora. We selected six populations from the Iberian collection of A. thaliana populations [11, 37,38,39,40,41] encompassing some of the environments where the species thrives across the region (Fig. 1 and Fig. S1). From a climatic viewpoint, populations represented mountain (AGU and CAI), continental (MAR and MDC) and coastal (BON and POB) Mediterranean climates. It must be noted that BON and POB, although both under a maritime influence, strongly differ in geographical location, altitude and distance to the coastline (Fig. 1 and Fig. S1). BON and CAI were the warmest and coolest populations, whereas MDC and CAI were the driest and the wettest populations, respectively (Fig. S1). Habitats included seaside stone pine (Pinus pinea L.) forests on sandy soils (BON), mixed forests dominated by oak species (POB), sclerophyllous scrublands with holm oak (Quercus ilex L.) trees (AGU, MAR and MDC), and mountainous Scots pine (Pinus sylvestris L.) forests (CAI) (Fig. S1).
Geographic location and vegetation type of A. thaliana populations with low (AGU, BON, and CAI) and high (MAR, MDC, and POB) genotypic and ecological diversity in edge and core environments, respectively. Upper panels depict the regional-scale proportion of suitable and unsuitable area (50 km radius) around each population estimated by pooling vegetation types with A. thaliana occurrences using the CORINE Land Cover 2000 database. Population maps are digitized aerial orthophotographs indicating vegetation types: forests (dark green), scrubland (light green), dry riverbeds (blue), boulders (grey), paths, and roads (orange). The location of sampled individuals for seeds and spots to estimate above and belowground abundance is indicated. Dot size is proportional to abundance. Scale bars indicate 100 m
Between early March and mid-June 2017, we haphazardly collected seeds from 50–60 individuals per population (area: 5.9–7.4 ha; Fig. 1 and Fig. S1). In the case of BON, we had to collect seed again in March 2018 to complete the panel of individuals. Maternal lines from BON generated from the 2018 sampling (see below) could not be used in analyses including the first experiment because they had not completed the after-ripening, affecting their performance in the first experiment (Supplementary Methods). We searched A. thaliana by covering sections within each population following a non-fixed zigzag path until completing the whole area [15]. In spring 2017 and 2018, we estimated aboveground abundance by haphazardly selecting 21–182 spots per population. (Fig. 1). In each spot, we counted A. thaliana individuals (all flowering and fruiting) in circular areas (5 m radius) with a hand tally counter.
By midsummer, A. thaliana seeds disperse and the first cm of soil are completely dry in all populations. Thus, in July 2018, we estimated belowground abundance (seed bank) by selecting 24 spots in each population based on the map of abundances (Fig. 1). In each spot, we collected soil from the upper soil layer (0–5 cm depth) at several points in circular areas (5 m radius). We pooled, sieved (to 2 mm) and stored dry soil samples from each spot in plastic tubes (225 ml). In October 2019, we conducted a germination experiment to estimate belowground A. thaliana abundance. We spread out soil samples (6 populations × 24 samples/population = 144 soil samples) on plastic trays (60 × 40 × 7 cm3) with moistened filter paper at the bottom. Plastic trays, covered with crystal-clear sealing film, were placed in a FITOCLIMA-10.000-EH growth chamber (ARALAB, Rio de Mouro, PT) at the Estación Biológica de Doñana (Sevilla, ES). We applied a cold treatment (4ºC in darkness over 2 weeks) to break seed dormancy, followed by a light treatment (20ºC in light over 1 week followed by 16 h of light/8 h of darkness over 3 additional weeks) to promote germination. We took pictures of each tray weekly and counted seedlings with the software ImageJ2 [42].
We used GPS (Garmin International, Inc., Olathe, US; positional error: 4 m) to record coordinates of individuals and spots. We calculated the proportion of each vegetation type in a circular area around the GPS coordinate of each individual (1 m radius) and spot (5 m radius) using a digitized aerial orthophotograph of each population [11, 15] from which vegetation type distributions were previously estimated (Fig. 1). The maximum membership proportion (range among populations = 0.91–0.98 and 0.68–0.79 for individuals and spots, respectively) was used to assign a vegetation type to each individual and spot. We computed the Shannon–Wiener index to estimate vegetation type diversity for individuals and spots.
Common garden experiments
Over fall and winter 2017–2018, we multiplied field-collected seeds via single-seed descent in a glasshouse at the Centro Nacional de Biotecnología (Madrid, ES). Overall, we obtained seeds from 55, 14, 50, 50, 53 and 53 maternal lines from AGU, BON, CAI, MAR, MDC and POB, respectively. In summer 2018, we multiplied seeds from 31 additional maternal lines collected in BON in March 2018 (Supplementary Methods), totaling 306 maternal lines. These materials are publicly available through the Nottingham Arabidopsis Stock Centre (NASC). We used multiplied seeds, minimizing environmental and maternal effects, to undertake common garden experiments at the Botanical Garden of Sierra de Grazalema Natural Park (36.46ºN, 5.30ºW; 329 m a.s.l.), over two years (2018–2019 and 2019–2020) to estimate variation in life-history traits and fitness [15, 39, 43, 44].
In summer 2018 and 2019, we prepared eight batches (replicates) of 60 seeds each per maternal line (306 maternal lines × 8 batches/maternal line × 60 seeds/batch = 146 880 seeds per experiment). We established experiments on the same date (November 15) and on the same exact stands to set up eight blocks (Fig. S2). We sowed seeds from each replicate and maternal line in square plastic pots (12 × 12 × 12 cm3) filled with standard soil mixture (Cejudo Baena S.L., Utrera, ES) and randomly placed one replicate per maternal line in each block (306 maternal lines × 8 replicates/maternal line = 2 448 pots per experiment).
We estimated recruitment as the maximum proportion of seedlings in each pot, recorded within the first 2–3 weeks after sowing. We estimated flowering time as the number of days between sowing and flowering dates, given by the date most plants in the pot (full-sibs with homogeneous behavior) had the first flower open. Paper bags were used to collect all plants when flowering finished and before fruit dehiscence. We recorded the number of plants per pot and the number of fruits per plant. Fecundity was estimated as the number of seeds per plant with a non-linear function relating the number of fruits per plant with the number of seeds per fruit [43]. We estimated survival as the proportion of plants relative to the maximum number of seedlings recorded. We estimated fitness (survival × fecundity) as the mean number of expected seeds per plant.
The national lockdown due to the coronavirus pandemic prevented us from monitoring the second experiment between mid-March and mid-June 2020. Nonetheless, we were able to impute flowering dates to those replicates of maternal lines with missing data. Practically all replicates of maternal lines from BON, MAR and MDC had complete flowering dates before the lockdown (Table S1), whereas 11, 21 and 38% of replicates of maternal lines from POB, CAI and AGU, respectively, had missing flowering dates. Imputation was feasible because we used the very same stands to set up the blocks in both experiments and because flowering dates were systematically earlier in the second experiment (Supplementary Methods; Table S1).
We recorded daily temperature and precipitation between establishment (November 15) and termination (April 15) dates in both experiments (Fig. S3). We obtained daily minimum and maximum temperature with HOBO Pendant UA-002–08 temperature loggers (Onset Computer Corporation, Inc., Bourne, US) and daily precipitation from data provided by the automatic meteorological station of the experimental facility.
Genetic characterization
We grew all A. thaliana maternal lines in a greenhouse at the Gregor Mendel Institute (Vienna Biocenter, Vienna, AT) to generate whole-genome shot-gun sequences (Supplementary Methods). Sequences were analyzed by the Service of Bioinformatics for Genomics and Proteomics at the Centro Nacional de Biotecnología (Madrid, ES). We generated a final VCF file containing 2 798 036 non-singleton nuclear SNPs genotyped in 298 of 306 maternal lines (Supplementary Methods). We generated a pairwise matrix of allele differences from this VCF file, which was used to identify pairs of maternal lines with nearly identical genotypes. Pairs of samples with genetic distances lower than 0.001 were considered as carrying the same genotype, because this was the genotyping error that we estimated by sequencing twice five MDC samples and in agreement with error sequencing rates described for Illumina short reads [45].
For each population, we assigned maternal lines to genotypes and computed the Shannon–Wiener index to estimate genotypic diversity. We did not detect shared genotypes among populations. As heterozygous SNPs were rescored to the major frequency allele to obtain the final VCF file (Supplementary Methods), we estimated mean observed heterozygosity per population with 12 neutral nuclear microsatellites genotyped in 282 of 306 maternal lines. To this end, we pooled DNA from six full-sibs from each maternal line grown in a growth chamber at the Estación Biológica de Doñana (Sevilla, ES) to extract DNA (details on DNA extraction, marker genotyping and genotyping error as in [37]).
Statistical analyses
We used linear mixed models to test the effect of group (fixed factor; populations with low and high genotypic and ecological diversity), experiment (fixed factor; 2018–2019 and 2019–2020), population nested within group (random factor; AGU, BON and CAI for the low diversity group in edge environments, and MAR, MDC and POB for the high diversity group in core environments), and maternal line (random factor) nested within population on life-history traits (recruitment, flowering time, survival and fecundity) and fitness in A. thaliana estimated in common garden experiments. The random interactions of experiment with population and maternal line were excluded from the model due to lack of convergence. As the model did not converge with survival, we replaced survival by the number of fruiting plants, used to estimate survival and closely related to recruitment (R2 = 0.71). We estimated variance components of random factors using the restricted maximum likelihood (REML) method. Significance of factors were tested using log-likelihood ratio tests and differences between population pairs were tested with Tukey post-hoc tests. We inspected variances of data, the existence of outliers and model residuals to check that the major assumptions of the analyses were acceptable. We fitted the linear mixed model using the MIXED procedure of SAS v.9.4 [46].
For each population and experiment, we estimated the correlation between pairs of traits with Dutilleul’s t-test, including the spatial autocorrelation of data, with the software SAM v.4.0 [47]. To estimate the contribution of genotypic variance to phenotypic variance of life-history traits and fitness in A. thaliana maternal lines from populations with low and high diversity in edge and core environments, respectively, estimated in the common garden experiments, we estimated broad-sense heritability (H2) values as H2 = VG/(VG + VE), where VG is the among-maternal line variance component and VE is the residual variance [48]. We estimated all variance components and their 95% confidence intervals using the remlVCA and VCAinference functions of the R package VCA v.1.4.3. (https://cran.rproject.org/web/packages/VCA/index.html).
We estimated phenotypic plasticity for life-history traits and fitness for each maternal line by computing the relative distance plasticity index (RDPI) [49], which ranges between 0 (no plasticity or canalization) and 1 (maximal plasticity or sensitivity), between the two common garden experiments. We tested the fixed effect of group (edge and core environments with low and high diversity, respectively) and the random effect of population nested within group on phenotypic plasticity of life-history traits and fitness with linear mixed models with the lmer function of the the R package lme4 [50] using maternal lines as replicates. We calculated the number of maternal lines from each population that significantly differed in life-history traits between experiments using Student’s t-tests. We also analyzed the distribution of maternal lines with low and high phenotypic plasticity among populations and traits with G-tests. To explore whether plasticity in recruitment and flowering time was related to fitness, we correlated plasticity distance matrices for these fitness components with fitness distance matrices with Mantel tests with PASSaGE v.2 [51].
We identified the drivers of variation in life-history traits and fitness of A. thaliana populations harboring low and high diversity in edge and core environments, respectively, by correlating pairwise phenotypic distance matrices with geographic, genetic and suitability distance matrices using Mantel tests. For each population, we obtained the geographic distance matrix as a matrix of Euclidian distances using GPS coordinates of sampled individuals. We generated the genetic distance matrix as a matrix of pairwise allelic differences among genotyped maternal lines with whole-genome sequences. We estimated a suitability distance matrix by using aboveground abundances, as spots with higher abundances are likely to be more suitable for A. thaliana. To do that, we generated a Voronoi diagram, in which each cell contained one spot to estimate aboveground abundance within each population (Fig. S4), with the function voronoi in the R package terra (https://cran.r-project.org/web/packages/terra/index.html). A suitability value to each sampled individual was assigned with the function st_intersection of the R package sf [52]. We performed all Mantel tests with standardized variables (subtracting the mean and scaling the variance) and estimated significances with 1000 permutations. Given the number of Mantel tests conducted, we only considered as significant those relationships with P < 0.01 (tests with P < 0.05 were few and with very low coefficients).
Finally, to identify the effects of selection on A. thaliana maternal lines from populations with low and high diversity in edge and core environments, respectively, estimated in the common garden experiments, we estimated linear and quadratic selection gradients (β and γ) and selection differentials (s and C) for recruitment and flowering time using traditional least squares-based regressions with fitness using the R package gsg [53]. We estimated all parameters from full models with linear and quadratic effects using standardized variables.
Results
Genetic and ecological attributes of study populations
The analysis of whole-genome sequences showed that the six A. thaliana study populations split into populations with low (AGU, BON and CAI) and high (MAR, MDC and POB) genotypic diversity in edge and core environments, respectively (Table 1). Populations with low genotypic diversity in edge environments had about one-fourth of maternal lines with different genotypes, of which about 50% or less were unique (represented by one maternal line only). In contrast, about four-fifths of maternal lines from populations with high genotypic diversity in core environments exhibited different genotypes, of which more than 80% were unique. Thus, the Shannon–Wiener index was lower in populations with low (range = 1.67–2.09) than high (range = 3.36–3.74) genotypic diversity. In addition, observed heterozygosity was 1–2 orders of magnitude lower in populations with low than high genotypic diversity in edge and core environments, respectively (Table 1).
From an ecological viewpoint, A. thaliana populations were located in contrasting environments chiefly defined by geography (Fig. 1), thereby affecting their climatic and ecological features (Fig. S1). The diversity of vegetation types within populations occupied by A. thaliana was the most remarkable difference between the two groups of populations (Table 1). Populations with low genotypic diversity in edge environments were massively dominated by forest and scrubland, whereas populations with high genotypic diversity in core environments encompassed a greater diversity of vegetation types, including forest, scrubland and openings made by dry riverbeds, boulders, or paths. The Shannon–Wiener index for vegetation type diversity of sampled individuals and spots to estimate abundances captured such differences: individuals and spots exhibited lower vegetation type diversity in populations with low (range = 0.54–0.76 and 0.66–0.85 for individuals and spots, respectively) than high (0.82–1.07 and 0.84–1.06 for individuals and spots, respectively) genotypic diversity in edge and core environments, respectively.
Above and belowground A. thaliana abundances (Fig. 1 and Fig. S4) did not differentiate populations with low and high diversity in edge and core environments, respectively. For example, the two populations with the highest mean aboveground abundance, but also with the highest variance, were AGU (low diversity) and POB (high diversity) (Table 1). The same applied for belowground abundance, as BON and AGU (both with low diversity) and MDC (high diversity) exhibited the highest mean belowground abundances (Table 1). Overall, there was no relationship between mean above and belowground abundances. For example, the two populations with the highest mean aboveground abundances (AGU and POB) had low and high mean belowground abundances (Table 1). Finally, BON was the population with the lowest mean aboveground abundances and had the highest mean belowground abundance, whereas CAI had the second lowest mean aboveground abundance and a nearly inexistent mean belowground abundance (Table 1).
Variation in life-history traits and fitness
We estimated the genetic component of variation in life-history traits and fitness of A. thaliana maternal lines from populations with low and high diversity in edge and core environments, respectively, in two common garden experiments. Weather conditions differed between experiments (Fig. S3). In particular, average minimum temperatures over the course of the first experiment were cooler (mean ± SD = 4.89 ± 2.85 ºC; range = -1.66–11.87 ºC) than those of the second experiment (6.44 ± 3.05 ºC; 1.46–14.65 ºC), although average maximum temperatures were similar in both experiments (23.47 ± 6.48 ºC and 23.14 ± 5.43 ºC for the first and second experiment, respectively). The second experiment was rainier (total rainfall over the experiment = 792.65 mm) than the first one (502.55 mm), although the amount of rainfall was more evenly distributed in the first (CV of rainfall over the experiment = 270.05%) than in the second (CV = 344.91%) experiment. On top of warmer temperatures, a heavy rain recorded in one day (180 mm on December 20, 2019) and a dry early spring with a delayed rainfall almost at the end of the experiment, affected the development of A. thaliana in the second experiment (Fig. S3), as shown by the number of maternal lines that did not complete the life cycle in the second experiment (range among populations = 2–15 maternal lines in POB and CAI, respectively).
Linear mixed models indicated that the two groups of populations with low and high diversity in edge and core environments, respectively, did not differ for recruitment, flowering time and survival (Table 2). In contrast, they significantly differed for fecundity and fitness (Table 2). Maternal lines from populations with low diversity produced less seeds per plant (mean ± SE = 111.25 ± 4.06 seeds per plant) than those with high diversity (144.11 ± 2.98 seeds per plant). Likewise, maternal lines from populations with low diversity exhibited lower fitness (78.98 ± 2.66 expected seeds per plant) than those with high diversity (102.87 ± 2.30 expected seeds per plant). In addition, all traits significantly differed between experiments (Table 2), although with different patterns. In particular, maternal lines showed higher recruitment (0.56 ± 0.01 and 0.47 ± 0.01 for the first and second experiment), later flowering time (115.81 ± 0.43 and 110.88 ± 0.55 days for the first and second experiment), higher survival (0.87 ± 0.01 and 0.57 ± 0.01 for the first and second experiment), and higher fitness (97.93 ± 2.19 and 86.26 ± 2.85 expected seeds/plant) in the first than in the second experiment. In contrast, maternal lines showed lower fecundity in the first (110.62 ± 2.30 seeds/plant) than in the second experiment (147.97 ± 4.27 seeds/plant).
Populations within groups (low and high diversity in edge and core environments, respectively) significantly differed for recruitment, flowering time and survival (Table 2 and Fig. 2), but with a different pattern. For example, for populations with low diversity in edge environments, BON was the only population significantly different from AGU and CAI for these three traits (|t|> 3.49, P < 0.007 in all cases; Tukey post-hoc test), whereas for populations with high diversity in core environments, POB was the only population significantly different from MAR and MDC for flowering time only (|t|> 5.19, P < 0.0001 in both cases; Tukey post-hoc test). Coefficients of variation for all traits also reflected these patterns of variation among populations within each diversity group (Table S2). The minimum and maximum coefficients of variation tended to be found among populations with low diversity (Table S2). Finally, maternal lines within each population exhibited significant differences for all traits (Table 2). Broad-sense heritability values were mostly significantly different from zero for recruitment and flowering time, whereas those for survival, fecundity and fitness tended to be indistinguishable from zero (Table S3), particularly in the first experiment.
Summary statistics for life-history traits and fitness of A. thaliana populations with low (AGU, BON, and CAI) and high (MAR, MDC, and POB) genotypic and ecological diversity in edge and core environments, respectively, estimated in two common garden experiments. For each population and experiment, boxes show the lower and upper quartiles, whiskers are drawn down to the 10th percentile and up to the 90th, the line is the median of observations, and dots indicate data points. Recruitment and survival are proportions, flowering time is given in days, and fecundity and fitness are given in seeds per plant and expected seeds per plant, respectively. Light and dark tones correspond to the first and second common garden experiments, respectively, whereas blue and ochre colors refer to populations from low and high diversity groups in edge and core environments, respectively
The pattern of pairwise correlation between traits for each population exhibited important differences between experiments (Table S4). In particular, significant correlations decreased in the second experiment (16 correlations) with respect to those detected in the first experiment (23 correlations). In general, populations with low diversity in edge environments showed a lower number of significant correlations between traits (17 correlations) than populations with high diversity in core environments (22 correlations). Overall, when significant, we observed a general trend: early flowering correlated with higher recruitment, higher survival, higher fecundity, and consequently higher fitness.
Phenotypic plasticity in life-history traits and fitness
Given the significant between-experiment differences for all traits, we estimated phenotypic plasticity between experiments for all maternal lines with complete data for all traits. Linear mixed models indicated that there were significant differences in phenotypic plasticity for recruitment, flowering time and fitness (λLR > 11.84, P < 0.004 in all cases; log-likelihood ratio tests), and non-significant for survival and fecundity (λLR < 0.01, P > 0.13 in all cases; log-likelihood ratio tests) (Fig. 3). Differences in phenotypic plasticity among populations within groups (low and high diversity in edge and core environments, respectively) tended to be more pronounced in populations with low than high diversity, particularly for recruitment, flowering time and fitness. Nevertheless, coefficients of variation for plasticity were rather similar among populations with minimum and maximum values detected in populations with either low or high diversity in edge and core environments, respectively (Table S2). The relationships between plasticity in fitness and plasticity in recruitment and flowering time were not significant in any population and experiment.
Summary statistics for plasticity of life-history traits and fitness of A. thaliana populations with low (AGU, BON, and CAI) and high (MAR, MDC, and POB) genotypic and ecological diversity in edge and core environments, respectively, estimated in two common garden experiments. For each population, boxes show the lower and upper quartiles, whiskers are drawn down to the 10th percentile and up to the 90th, the line is the median of observations, and dots indicate data points. For each trait, different letters indicate significance differences among populations within each diversity group (P < 0.05; Tukey post-hoc tests). Light and dark green correspond to populations from low and high diversity groups in edge and core environments, respectively
All populations showed maternal lines with low and high plasticity, as indicated by the number of maternal lines per population falling in the upper and lower 10th percentiles of phenotypic variation (Table 3). The number of maternal lines across populations and traits was unevenly distributed for the upper 10th percentile of phenotypic plasticity (χ2 = 69.36, P < 0.0001; G-test), but not for the lower 10th percentile (χ2 = 31.06, P = 0.054; G-test). For the upper 10th percentile, we found a high number of plastic maternal lines for recruitment in CAI (low diversity) and for flowering time in MDC (high diversity), and a lack of plastic maternal lines for recruitment and survival in BON (low diversity) and for recruitment and fecundity in POB (high diversity).
Drivers of phenotypic variation and plasticity in life-history traits and fitness
We explored the effects of ecological and genetic drivers of variation on life-history traits and fitness of maternal lines from populations with low and high diversity in edge and core environments, respectively (Table 4). The results indicated that, when significant, genetic similarity (14 cases across all populations, except BON) was more important than geographic similarity (4 cases in CAI) for trait differentiation. We detected more significant relationships in the first (11 cases in flowering time, fecundity and fitness) than in the second (7 cases time in flowering and survival) experiment. Flowering time was the trait with the highest number of significant correlations (11 cases), followed by fitness (3 cases), fecundity (3 cases) and survival (1 case).
Correlations between phenotypic distance and suitability distance were only significant for flowering time in CAI (low diversity; r = 0.38, P = 0.001; second experiment; Mantel test) and POB (high diversity; r = 0.48, P = 0.001; first experiment; Mantel test). Correlations between phenotypic plasticity distance and geographic distance were only significant for recruitment in AGU (low diversity; r = 0.30, P = 0.001; Mantel test) and fitness in BON (low diversity; r = 0.35, P = 0.007; Mantel test), whereas none of the correlations between phenotypic plasticity distance and genetic or suitability distance were significant (range of |r|= 0.00–0.29, P > 0.011 in all cases; Mantel tests).
Selection analysis
We estimated selection gradients and selection differentials for recruitment and flowering time to examine how natural selection acted upon A. thaliana phenotypes in the common garden experiments. Linear selection gradients were significant and negative for flowering time in the first experiment in all populations (Table 5 and Table S5), indicating that selection favored A. thaliana individuals with earlier flowering in the conditions encountered in the first experiment. In the second experiment, significant linear selection gradients were scarce, weaker and very different from those detected in the first experiment (Table 5 and Table S5). In particular, we found a significant negative linear selection gradient for recruitment in CAI (low diversity), suggesting that selection favored A. thaliana individuals with lower recruitment, and a significant positive linear selection gradient for flowering time in POB (high diversity), suggesting that selection favored A. thaliana individuals with later flowering.
Variation in life-history traits and fitness at the maternal line level
We tested the differences between experiments in life-history traits for each maternal line from populations with low and high diversity. The results indicated that all populations, irrespective of their environment type with distinct genotypic and ecological diversity, exhibited maternal lines with significant and non-significant variation between experiments in all traits (Fig. 4). Although this experiment was not conceived to quantify differences among maternal lines within genotypes, we detected maternal lines sharing the same (nearly identical) genotype with significant and non-significant differences between experiments for almost all traits in populations with low and high diversity in edge and core environments, respectively (Fig. S5).
Reaction norms of maternal lines from A. thaliana populations with low (AGU, BON, and CAI) and high (MAR, MDC, and POB) genotypic and ecological diversity in edge and core environments, respectively, with significant (red lines: decrease; black lines: increase) and non-significant (grey lines) differences in life-history traits and fitness between the two common garden experiments
In BON (low diversity), all maternal lines with significant differences between experiments exhibited lower values for all traits in the second experiment (Fig. 4). There were generalized decreases in the second experiment for recruitment, flowering time and survival for all populations, except one individual from MDC and one from POB (both with high diversity) that increased recruitment in the second experiment (Fig. 4). In contrast, fecundity and fitness exhibited further variability (Fig. 4). Maternal lines with significant differences between experiments increased fecundity in the second experiment in two populations with low (AGU and CAI) and high (MDC and POB) diversity. In MAR (high diversity), there were two maternal lines that increased and two that decreased fecundity in the second experiment. In the case of fitness, all maternal lines with significant differences between experiments decreased fitness in the second experiment in all populations with high diversity (Fig. 4). In contrast, in CAI (low diversity) there was one maternal line that increased fitness in the second experiment, and AGU (low diversity) exhibited four and three maternal lines that increased and decreased fitness in the second experiment, respectively (Fig. 4).
The number of maternal lines with significant variation between experiments was unevenly distributed across populations and traits (χ2 = 73.13, P < 0.0001; G-test). However, there were differences between groups of populations. Populations with low diversity in edge environments significantly differed among them in the distribution of maternal lines with significant variation in traits between experiments, particularly for the comparisons between BON and AGU (χ2 = 13.21, P = 0.004; G-test), and BON and CAI (χ2 = 10.95, P = 0.012; G-test). In contrast, all populations with high diversity in core environments did not show significant differences among them in the distribution of maternal lines with significant variation in traits between experiments (χ2 < 4.94, P > 0.18 in all cases; G-tests).
Discussion
The amount of genetic and phenotypic variation that a single population can harbor and the relationship between these two structural components of populations are far from being understood due to the intertwined interaction among genetic differences, environmental influences and stochastic events affecting the genotype–phenotype correspondence [54]. Here, we tackled this question by estimating variation in life-history traits and fitness of A. thaliana maternal lines from Iberian populations characterized by low and high genotypic and ecological diversity in edge and core environments, respectively. Although the large-scale effects of environmental and ecological variation on A. thaliana’s life-history traits are well known [23, 38, 55,56,57,58], the impact of environmental variation on within-population genetic and phenotypic variation is less understood [11, 15, 30, 44, 47, 59,60,61], mainly because working with populations is far more challenging and demanding than handling accessions. Hence, population-based approaches represent a fundamental piece to better grasp the species’ evolutionary ecology and its response to environmental changes.
In this study, we first expected that A. thaliana populations from edge environments with low genotypic and ecological diversity would exhibit lower phenotypic variation, as directional selection in edge environments should have imposed stronger shifts in trait distribution by increasing the frequency of genotypes with eventually higher fitness [57, 62]. This process may occur particularly fast in a scenario of restricted gene flow [63, 64], such as Iberian A. thaliana [11]. The results, however, showed that all populations expressed a substantial amount of phenotypic variation in practically all traits in our common garden experiments (Fig. 2 and Table S2). In addition, we found consistent patterns of variation for recruitment and flowering time, as these two traits did not differ between groups of populations, had non-zero broad-sense heritability values in almost all populations and experiments (Table S3), and represented major targets of directional selection, particularly flowering time in the first experiment (Table 5 and Table S5). Hence, selection has probably operated in all populations by adjusting the sequential nature of germination and flowering phenology to their own environments [65,66,67], but without depleting phenotypic variability, as observed in other annuals even in extremely harsh environments [68]. A recent study on phenotypic variation in urban A. thaliana populations also indicated that environmental filtering in cities promoted the maintenance of phenotypic diversity in fitness-related traits [69], stressing the role of environmental heterogeneity for the preservation of genetic diversity in plants.
Based on the dramatic change in the intensity and direction of selection on flowering time between experiments, as observed elsewhere in A. thaliana [43, 67, 70,71,72], and to a much lesser extent on recruitment in the second experiment (Table 5), we believe that year-to-year fluctuations in environmental-driven selection is one of the forces accounting for the maintenance of phenotypic variation in populations. In fact, former studies showed that the consequences for life-history traits in populations with fluctuating and episodic selection pressures are highly context-dependent, leading to fluctuating selection responses and the maintenance of genetic variation within populations [73, 74], likely contributing to the evolutionary success of populations.
On top of that, recent findings on the extent of local adaptation using long-term experiments with recombinant inbred lines in A. thaliana, indicated that even locally adapted populations may not reach their fitness optimum in their environments due to the presence of maladaptive loci that remain in populations [75]. Furthermore, a limited number of genetic trade-offs and conditionally adaptive loci emerge as responsible for local adaptation in A. thaliana [75, 76] as well as in other plants [77,78,79,80,81]. As the eventual effects of genetic trade-offs and conditional neutrality depend on the environmental context and are subject to temporally variable selection [75], A. thaliana populations possess the means to maintain phenotypic variation in the long run. Despite the low gene flow and low outcrossing rates in natural A. thaliana populations [11] and the contribution of mutation accumulation to standing genetic variation and phenotypic variation [82], every effective recombination between two genetically and phenotypically distinct individuals suffices to yield rapid changes in phenotypic variation [83] and has the potential to create a progeny with a considerable amount of variation and plasticity in fitness-related traits, such as flowering time [84]. This property also accounts for the great amount of phenotypic variation and plasticity detected in our A. thaliana populations with either low or high genotypic and ecological diversity.
However, we detected slight differences between A. thaliana populations with low and high diversity that are worth mentioning. For example, populations with low diversity exhibited lower fecundity and fitness, traits whose variance is mostly determined by its environmental component, than populations with high diversity (Fig. 2). This result suggests that harboring low genotypic diversity might entail a stronger, albeit not critical, response of populations to environmental changes in an important fitness component, such as fecundity. Nonetheless, A. thaliana just needs a favorable year to boost fecundity to replenish the soil seed bank and potentially increase plant density in forthcoming years [15, 18], which may easily buffer the detrimental effects of poor years on fecundity. The long-term monitoring of one of our study populations since 2012 (MDC) supports this view [15], as it exhibited large year-to-year fluctuations in abundance but with no effects on the genetic and phenotypic composition. The dramatic changes in abundance in A. thaliana populations over time might account for the lack of a clear relationship between above and belowground abundance, as well as between abundance and genotypic diversity in our study populations. Furthermore, BON significantly differed from the other two populations with low diversity (AGU and CAI) for recruitment, flowering time and survival. In contrast, POB significantly differed from the other two populations with high diversity (MAR and MDC) for flowering time only. A similar picture was detected for among-population variation in phenotypic plasticity, as BON and POB stood out as the two populations differing from the other populations in their diversity groups (Fig. 3). Thus, local environmental features, irrespective of genotypic diversity, may strongly affect fitness-related traits and their plasticity, as BON (a seaside population) differed from AGU and CAI (two mountain pass populations), whilst POB (a coastal mixed forest population) differed from MAR and MDC (two continental scrubland populations) in several traits.
Our second expectation dealt with the effects of genotypic and ecological diversity on phenotypic plasticity of A. thaliana traits, which was supported by the positive relationship between environmental heterogeneity and phenotypic plasticity found in other plants [31, 32, 85, 86]. Our common garden experiments, carried out under unplanned benign and harsh conditions for A. thaliana, allowed us to estimate the response of maternal lines from each population to warmer temperatures and more seasonal precipitation (Fig. S3), which remarkably mimicked the predicted climatic trend across the Mediterranean Basin region in the near future [87]. Regardless of their genotypic and ecological diversity, we found that all populations encompassed maternal lines with low and high levels of phenotypic plasticity with significant differences between experiments in practically all populations and traits (Fig. 4 and Table 3). Other studies on phenotypic plasticity in A. thaliana also indicated that all life stages with no exception may exhibit a plastic behavior [88]. The potential shown by maternal lines from any population to be plastic for almost any trait aligns with field observations [89], experiments [65, 90] and models [91] indicating that a single genotype has the ability to produce mixtures of phenotypes differing in traits related to life-cycle variation in A. thaliana.
Given the lack of a relationship between phenotypic plasticity and fitness, we suggest a non-adaptive nature of phenotypic plasticity in this set of Iberian A. thaliana populations, as also found in experiments using genetic constructs [92, 93] and populations from the non-native North American range [30]. In contrast, regional-scale studies using accessions did find signs of adaptive plasticity in A. thaliana, either by significant relationships with fitness [43] or with the environmental variability of origin [94], suggesting that scale matters in depicting the adaptive significance of phenotypic plasticity in A. thaliana. Once more, the patterns that clearly emerge with accession-based approaches at a regional scale are much more difficult to detect at the population scale.
Despite the existence of maternal lines with significant variation between experiments in all populations (Fig. 4), mostly decreasing life-history traits and fitness, canalized maternal lines with non-significant differences between experiments were substantial or even dominant in all populations. We estimated that between 40 and 80% of maternal lines that completed the life cycle in the second experiment exhibited canalization across populations and traits, irrespective of their diversity group. The duality of maternal lines with a trend for plasticity and canalization coexisting within the same population is supported by theoretical work indicating that plasticity and canalization in populations are favored in epochs of environmental shifts and stability, respectively [95]. Nevertheless, the genetic and epigenetic contributions to integrated phenotypes [96] along with the degree of environmental fluctuations may determine the extent of plasticity and canalization. In fact, in A. thaliana, we know that specific allele combinations of seed dormancy and/or flowering time genes determine key developmental traits as well as their phenotypic plasticity [84, 92, 93, 97, 98] and that the diversity of parental environments modulates phenotypic modifications in the offspring for a few generations [35, 88, 99] through environmentally-induced epigenetic changes [100, 101].
Conclusions
Natural A. thaliana populations that have remained undisturbed for long time are able to maintain phenotypic variation and phenotypic plasticity for life-history traits with an important contribution to fitness. Edge and core environments with low and high ecological diversity, respectively, may influence the genotypic composition of A. thaliana populations, probably through effective recombination from very different outcrossing rates between edge and core environments (Table 1), but seem not to determine their phenotypic diversity in the response of genotypes to the idiosyncrasy of each environment. Beyond the level of genetic variation harbored by a population, maintaining canalized and sensitive phenotypes to environmental fluctuations within populations represents an extraordinary guarantee to endure in changing environments, but only demanding within-population approaches have the power to depict such patterns.
Availability of data and materials
Data deposited in the Dryad repository: https://doi.org/10.5061/dryad.98sf7m0qp [102]. Seeds are publicly available through the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info). Raw whole-genome sequences are available at NCBI SRA (BioProject number PRJNA998580; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA998580). Field sampling was conducted in locations where permission was not required, except at BON that is located within Doñana Natural Area (permission issued by Estación Biológica de Doñana on March 3, 2016; Ref.: 20151073000000570). Based on the Royal Decree of the Spanish legislation (Real Decreto 124/2017, de 24 de febrero; https://www.boe.es/eli/es/rd/2017/02/24/124), the genetic resources included in this study fall within the definition of “taxonomic purposes”.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- GPS:
-
Global positioning system
- RDPI:
-
Relative distance plasticity index
- REML:
-
Restricted maximum likelihood
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeats
- VCF:
-
Variant call format
- WGS:
-
Whole-genome sequence
References
Carley LN, Morris WF, Walsh R, Riebe D, Mitchell-Olds T. Are genetic variation and demographic performance linked? Evol Appl. 2022;15:1888–906. https://doi.org/10.1111/eva.13487.
Chevin LM, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: Towards a predictive theory. PLoS Biol. 2010;8:e1000357. https://doi.org/10.1371/journal.pbio.1000357.
Frankham R. Relationship of genetic variation to population size in wildlife. Conserv Biol. 1996;10:1500–8. https://doi.org/10.1046/j.1523-1739.1996.10061500.x.
Leimu R, Mutikainen P, Koricheva J, Fischer M. How general are positive relationships between plant population size, fitness and genetic variation? J Ecol. 2006;94:942–52. https://doi.org/10.1111/j.1365-2745.2006.01150.x.
Butlin RK, Tregenza T. Levels of genetic polymorphism: marker loci versus quantitative traits. Phil Trans R Soc Lond B. 1998;353:187–98. https://doi.org/10.1098/2Frstb.1998.0201.
Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis Evolution. 2001;55:1095–103. https://doi.org/10.1111/j.0014-3820.2001.tb00629.x.
McKay JK, Latta RG. Adaptive population divergence: markers QTL and traits. Trends Ecol Evol. 2002;17:285–91. https://doi.org/10.1016/S0169-5347(02)02478-3.
Villellas J, Berjano R, Terrab A, Garcia MB. Divergence between phenotypic and genetic variation within populations of a common herb across Europe. Ecosphere. 2014;5:56. https://doi.org/10.1890/ES13-00291.1.
Volis S, Ormanbekova D, Yermekbayev K, Song M, Shulgina I. The conservation value of peripheral populations and a relationship between quantitative trait and molecular variation. Evol Biol. 2016;43:26–36. https://doi.org/10.1007/s11692-015-9346-3.
Walisch TJ, Colling G, Hermant S, Matthies D. Molecular and quantitative genetic variation within and between populations of the declining grassland species Saxifraga granulata. Ecol Evol. 2022;12:e9462. https://doi.org/10.1002/ece3.9462.
Castilla AR, Méndez-Vigo B, Marcer A, Martínez-Minaya J, Conesa D, Picó FX, et al. Ecological, genetic and evolutionary drivers of regional genetic differentiation in Arabidopsis thaliana. BMC Evol Biol. 2020;20:71. https://doi.org/10.1186/s12862-020-01635-2.
Lundemo S, Falahati-Anbaran M, Stenøien HK. Seed banks cause elevated generation times and effective population sizes of Arabidopsis thaliana in northern Europe. Mol Ecol. 2009;18:2798–811. https://doi.org/10.1111/j.1365-294x.2009.04236.x.
Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, et al. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 2010;6:e1000890. https://doi.org/10.1371/journal.pgen.1000890.
Falahati-Anbaran M, Lundemo S, Stenøien HK. Seed dispersal in time can counteract the effect of gene flow between natural populations of Arabidopsis thaliana. New Phytol. 2014;202:1043–54. https://doi.org/10.1111/nph.12702.
Méndez-Vigo B, Castilla AR, Gómez R, Marcer A, Alonso-Blanco C, Picó FX. Spatiotemporal dynamics of genetic variation at the quantitative and molecular levels within a natural Arabidopsis thaliana population. J Ecol. 2022;110:2701–16. https://doi.org/10.1111/1365-2745.13981.
Galloway LF. Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytol. 2005;166:93–100. https://doi.org/10.1111/j.1469-8137.2004.01314.x.
Chevin LM, Lande R. Adaptation to marginal habitats by evolution of increased phenotypic plasticity. J Evol Biol. 2011;24:1462–76. https://doi.org/10.1111/j.1420-9101.2011.02279.x.
Matesanz S, Ramírez-Valiente JA. A review and meta-analysis of intraspecific differences in phenotypic plasticity: Implications to forecast plant responses to climate change. Global Ecol Biogeogr. 2019;28:1682–94. https://doi.org/10.1111/geb.12972.
Pironon S, Papuga G, Villellas J, Angert AL, García MB, Thompson JD. Geographic variation in genetic and demographic performance: new insights from an old biogeographical paradigm. Biol Rev Camb Philos Soc. 2017;92:1877–909. https://doi.org/10.1111/brv.12313.
Bontrager M, Usui T, Lee-Yaw JA, Anstett DN, Branch HA, Hargreaves AL, et al. Adaptation across geographic ranges is consistent with strong selection in marginal climates and legacies of range expansion. Evolution. 2021;75:1316–33. https://doi.org/10.1111/evo.14231.
Gomaa NH, Montesinos-Navarro A, Alonso-Blanco C, Picó FX. Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol Ecol. 2011;20:3540–54. https://doi.org/10.1111/j.1365-294x.2011.05193.x.
Montesinos-Navarro A, Wig J, Picó FX, Tonsor ST. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 2011;189:282–94. https://doi.org/10.1111/j.1469-8137.2010.03479.x.
Vidigal DS, Marques ACSS, Willems LAJ, Buijs G, Méndez-Vigo M, Hilhorst HWM, et al. Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ. 2016;39:1737–48. https://doi.org/10.1111/pce.12734.
Picó FX. Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. J Ecol. 2012;100:1009–18. https://doi.org/10.1111/j.1365-2745.2012.01979.x.
Argyres AZ, Schmitt J. Microgeographic genetic structure of morphological and life history traits in a natural population of Impatiens capensis. Evolution. 1991;45:175–89. https://doi.org/10.1111/j.1558-5646.1991.tb05276.x.
Galloway LF. Response to natural environmental heterogeneity: Maternal effects and selection on life-history characters and plasticities in Mimulus guttatus. Evolution. 1995;49:1095–107. https://doi.org/10.2307/2410434.
Prati D, Schmid B. Genetic differentiation of life-history traits within populations of the clonal plant Ranunculus reptans. Oikos. 2000;90:442–56. https://doi.org/10.1034/j.1600-0706.2000.900303.x.
Weinig C, Dorn LA, Kane NC, German ZM, Halldorsdottir SS, Ungerer MC, et al. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics. 2003;165:321–9. https://doi.org/10.1093/genetics/165.1.321.
Paccard A, Vance M, Willi Y. Weak impact of fine-scale landscape heterogeneity on evolutionary potential in Arabidopsis lyrata. J Evol Biol. 2013;26:2331–40. https://doi.org/10.1111/jeb.12220.
Callahan HS, Pigliucci M. Shade-induced plasticity and its ecological significance in wild populations of Arabidopsis thaliana. Ecology. 2002;83:1965–80. https://doi.org/10.2307/3071779.
Lázaro-Nogal A, Matesanz S, Godoy A, Pérez-Trautman F, Gianoli E, Valladares F. Environmental heterogeneity leads to higher plasticity in dry-edge populations of a semi-arid Chilean shrub: insights into climate change responses. J Ecol. 2015;103:338–50. https://doi.org/10.1111/1365-2745.12372.
de la Mata R, Zas R, Bustingorri G, Sampedro L, Rust M, Hernandez-Serrano A, et al. Drivers of population differentiation in phenotypic plasticity in a temperate conifer: A 27-year study. Evol Appl. 2022;15:1945–62. https://doi.org/10.1111/eva.13492.
Galloway LF, Etterson JR. Transgenerational plasticity is adaptive in the wild. Science. 2007;318:1134–6. https://doi.org/10.1126/science.1148766.
Auge GA, Leverett LD, Edwards BR, Donohue K. Adjusting phenotypes via within- and across-generational plasticity. New Phytol. 2017;216:343–9. https://doi.org/10.1111/nph.14495.
Schmid MW, Heichinger C, Coman Schmid D, Guthörl D, Gagliardini V, Bruggmann R, et al. Contribution of epigenetic variation to adaptation in Arabidopsis. Nat Commun. 2018;9:4446. https://doi.org/10.1038/s41467-018-06932-5.
Yan Z, Tian D, Han W, Ji C, Hou X, Guo Y, et al. Weak transgenerational effects of ancestral nitrogen and phosphorus availabilities on offspring phenotypes in Arabidopsis thaliana. J Plant Res. 2023;136:515–25. https://doi.org/10.1007/s10265-023-01456-6.
Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian Peninsula. Genetics. 2008;180:1009–21. https://doi.org/10.1534/genetics.108.089581.
Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC and PHYC genes in Arabidopsis. Plant Physiol. 2011;157:1942–55. https://doi.org/10.1104/pp.111.183426.
Manzano-Piedras E, Marcer A, Alonso-Blanco C, Picó FX. Deciphering the adjustment between environment and life history in annuals: lessons from a geographically-explicit approach in Arabidopsis thaliana. PLoS ONE. 2014;9:e87836. https://doi.org/10.1371/journal.pone.0087836.
Marcer A, Vidigal DS, James PMA, Fortin MJ, Méndez-Vigo B, Hilhorst HWM, et al. Temperature fine-tunes Mediterranean Arabidopsis thaliana life-cycle phenology geographically. Plant Biol. 2018;20:148–56. https://doi.org/10.1111/plb.12558.
Tabas-Madrid D, Méndez-Vigo B, Arteaga N, Marcer A, Pascual-Montano A, Weigel D, et al. Genome-wide signatures of flowering adaptation to climate temperature: Regional analyses in a highly diverse native range of Arabidopsis thaliana. Plant Cell Environ. 2018;41:1806–20. https://doi.org/10.1111/pce.13189.
Rueden C, Schindelin TJ, Hiner MC, DeZonia BE, Walter AE, Arena ET, et al. Image J2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. https://doi.org/10.1186/s12859-017-1934-z.
Exposito-Alonso M, Brennan AC, Alonso-Blanco C, Picó FX. Spatio-temporal variation in fitness responses to contrasting environments in Arabidopsis thaliana. Evolution. 2018;72:1570–86. https://doi.org/10.1111/evo.13508.
Gómez R, Méndez-Vigo B, Marcer A, Alonso-Blanco C, Picó FX. Quantifying temporal change in plant population attributes: insights from a resurrection approach. AoB Plants. 2018;10:ply063. https://doi.org/10.1093/aobpla/ply063.
Stoler N, Nekrutenko A. Sequencing error profiles of Illumina sequencing instruments. NAR Genom Bioinform. 2021;27:lqad019. https://doi.org/10.1093/nargab/lqab019.
SAS. Statistical analysis software. Users’ guide statistics version 9.4. Cary: SAS Institute Inc.; 2013.
Rangel TF, Diniz-Filho JAF, Bini LM. SAM: a comprehensive application for spatial analysis in macroecology. Ecography. 2010;33:46–50. https://doi.org/10.1111/j.1600-0587.2009.06299.x.
Le Corre V. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol Ecol. 2005;14:4181–92. https://doi.org/10.1111/j.1365-294x.2005.02722.x.
Valladares F, Sánchez-Gómez D, Zabala MA. Quantitative estimation of phenotypic plasticity: bridging the gap between the evolutionary concept and its ecological applications. J Ecol. 2006;94:1103–16. https://doi.org/10.1111/j.1365-2745.2006.01176.x.
Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. https://doi.org/10.18637/jss.v067.i01.
Rosenberg MS, Anderson CD. PASSaGE: Pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol Evol. 2011;2:229–32. https://doi.org/10.1111/j.2041-210X.2010.00081.x.
Pebesma E. Simple features for R Standardized support for spatial vector data. The R J. 2018;10:439–46. https://journal.r-project.org/archive/2018/RJ-2018-009/index.html.
Morrissey MB, Sakrejda K. Unification of regression-based methods for the analysis of natural selection. Evolution. 2013;67:2094–100. https://doi.org/10.1111/evo.12077.
Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJ, Schubart CD. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol. 2008;211:510–23. https://doi.org/10.1242/jeb.008755.
Debieu M, Tang C, Stich B, Sikosek T, Effgen S, Josephs E, et al. Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS ONE. 2013;8:e61075. https://doi.org/10.1371/journal.pone.0061075.
Vasseur F, Exposito-Alonso M, Ayala-Garay OJ, Wang G, Enquist BJ, Vile D, et al. Adaptive diversification of growth allometry in the plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 2018;115:3416–21. https://doi.org/10.1073/pnas.1709141115.
Exposito-Alonso M, 500 Genomes Field Experiment Team, Burbano HA, Bossdorf O, Nielsen R, Weigel D. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature. 2019;573:126–9. https://doi.org/10.1038/s41586-019-1520-9.
Martínez-Berdeja A, Stitzer MC, Taylor MA, Okada M, Ezcurra E, Runcie DE, et al. Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2020;117:2526–34. https://doi.org/10.1073/pnas.1912451117.
Stenøien HK, Fenster CB, Tonteri A, Savolainen O. Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Mol Ecol. 2005;14:137–48. https://doi.org/10.1111/j.1365-294x.2004.02359.x.
Frachon L, Libourel C, Villoutreix R, Carrère S, Glorieux C, Huard-Chauveau C, et al. Intermediate degrees of synergistic pleiotropy drive adaptive evolution in ecological time. Nat Eco Evol. 2017;1:1551–61. https://doi.org/10.1038/s41559-017-0297-1.
Murren CJ, Alt CHS, Kohler C, Sancho G. Natural variation on whole-plant form in the wild is influenced by multivariate soil nutrient characteristics: natural selection acts on root traits. Am J Bot. 2020;107:319–28. https://doi.org/10.1002/ajb2.1420.
Hoffmann AA, Hercus MJ. Environmental stress as an evolutionary force. Bioscience. 2000;50:217–26. https://doi.org/10.1641/0006-3568(2000)050[0217:ESAAEF]2.3.CO;2.
García-Ramos G, Kirkpatrick M. Genetic models of adaptation and gene flow in peripheral populations. Evolution. 1997;51:21–8. https://doi.org/10.1111/j.1558-5646.1997.tb02384.x-.
Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–41. https://doi.org/10.1111/j.1461-0248.2004.00684.x.
Donohue K. Completing the cycle: maternal effects as the missing link in plant life histories. Phil Trans R Soc Lond B. 2009;364:1059–74. https://doi.org/10.1098/rstb.2008.0291.
Galloway LF, Burgess KS. Manipulation of flowering time: Phenological integration and maternal effects. Ecology. 2009;90:2139–48. https://doi.org/10.1890/08-0948.1.
Taylor MA, Cooper MD, Sellamuthu R, Braun P, Migneault A, Browning A, et al. Interacting effects of genetic variation for seed dormancy and flowering time on phenology, life history, and fitness of experimental Arabidopsis thaliana populations over multiple generations in the field. New Phytol. 2017;216:291–302. https://doi.org/10.1111/nph.14712.
Gomaa NH, Picó FX. Depicting the phenotypic space of the annual plant Diplotaxis acris in hyperarid deserts. Ecol Evol. 2021;11:15708–19. https://doi.org/10.1002/ece3.8232.
Schmitz G, Linstädter A, Frank ASK, Dittberner H, Thome J, Schrader A, et al. Environmental filtering of life-history trait diversity in urban populations of Arabidopsis thaliana. J Ecol. 2023;112:12–27. https://doi.org/10.1111/1365-2745.14211.
Fournier-Level A, Wilczek AM, Cooper MD, Roe JL, Anderson J, Eaton D, et al. Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol Ecol. 2013;22:3552–66. https://doi.org/10.1111/mec.12285.
Ågren J, Oakley CG, Lundemo S, Schemske DW. Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: Estimates of selection and QTL mapping. Evolution. 2017;71:550–64. https://doi.org/10.1111/evo.13126.
Postma FM, Ågren J. Among-year variation in selection during early life stages and the genetic basis of fitness in Arabidopsis thaliana. Mol Ecol. 2018;27:2498–511. https://doi.org/10.1111/mec.14697.
Siepielski AM, DiBattista JD, Carlson SM. It’s about time: the temporal dynamics of phenotypic selection in the wild. Ecol Lett. 2009;12:1261–76. https://doi.org/10.1111/j.1461-0248.2009.01381.x.
Monro K, Marshall DJ. Faster is not always better: selection on growth rate fluctuates across life history and environments. Am Nat. 2014;183:798–809. https://doi.org/10.1086/676006.
Oakley CG, Schemske DW, McKay JK, Ågren J. Ecological genetics of local adaptation in Arabidopsis: An 8-year field experiment. Mol Ecol. 2023;32:4570–83. https://doi.org/10.1111/mec.17045.
Ågren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2013;110:21077–82. https://doi.org/10.1073/pnas.1316773110.
Lowry DB, Hall MC, Salt DE, Willis JH. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytol. 2009;183:776–88. https://doi.org/10.1111/j.1469-8137.2009.02901.x.
Anderson JT, Lee CR, Rushworth CA, Colautti RI, Mitchell-Olds T. Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol. 2013;22:699–708. https://doi.org/10.1111/j.1365-294x.2012.05522.x.
Leinonen PH, Remmington DL, Leppälä J, Savolainen O. Genetic basis of local adaptation and flowering time variation in Arabidopsis lyrata. Mol Ecol. 2013;22:709–23. https://doi.org/10.1111/j.1365-294x.2012.05678.x.
Wadgymar SM, Lowry DB, Gould BA, Byron CN, Mactavish RM, Anderson JT. Identifying targets and agents of selection: Innovative methods to evaluate the processes that contribute to local adaptation. Methods Ecol Evol. 2017;8:738–49. https://doi.org/10.1111/2041-210X.12777.
Wright SJ, Goad DM, Gross BL, Muñoz PR, Olsen KM. Genetic trade-offs underlie divergent life history strategies for local adaptation in white clover. Mol Ecol. 2021;31:3742–60. https://doi.org/10.1111/mec.16180.
Rutter MT, Roles AJ, Fenster CB. Quantifying natural seasonal variation in mutation parameters with mutation accumulation lines. Ecol Evol. 2018;8:5575–85. https://doi.org/10.1002/ece3.4085.
Palacio-Lopez K, Molofsky J. Phenotypic shifts following admixture in recombinant offspring of Arabidopsis thaliana. Evol Ecol. 2021;35:575–93. https://doi.org/10.1007/s10682-021-10118-9.
Méndez-Vigo B, Savic M, Ausín I, Ramiro M, Martín B, Picó FX, et al. Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant Cell Environ. 2016;39:282–94. https://doi.org/10.1111/pce.12608.
Gianoli E, González-Teuber M. Environmental heterogeneity and population differentiation in plasticity to drought in Convolvulus chilensis (Convolvulaceae). Evol Ecol. 2005;19:603–13. https://doi.org/10.1007/s10682-005-2220-5.
Molina-Montenegro MA, Atala C, Gianoli E. Phenotypic plasticity and performance of Taraxacum officinale (dandelion) in habitats of contrasting environmental heterogeneity. Biol Invasions. 2010;12:2277–84. https://doi.org/10.1007/s10530-009-9638-6.
Gómez-Navarro JJ, Montávez JP, Jiménez-Guerrero P, Jerez S, García-Valero JA, González-Rouco JF. Warming patterns in regional climate change projections over the Iberian Peninsula. Meteorol Z. 2010;19:275–85. https://doi.org/10.1127/0941-2948/2010/0351.
Alvarez M, Bleich A, Donohue K. Genotypic variation in the persistence of transgenerational responses to seasonal cues. Evolution. 2020;74:2265–80. https://doi.org/10.1111/evo.13996.
Montesinos A, Tonsor ST, Alonso-Blanco C, Picó FX. Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE. 2009;4:e7213. https://doi.org/10.1371/journal.pone.0007213.
Montesinos-Navarro A, Picó FX, Tonsor SJ. Clinal variation in seed traits influencing life cycle timing in Arabidopsis thaliana. Evolution. 2012;66:3417–31. https://doi.org/10.1111/j.1558-5646.2012.01689.x.
Burghardt LT, Jessica C, Metcalf E, Wilczek AM, Schmitt J, Donohue K. Modeling the influence of genetic and environmental variation on the expression of plant life cycles across landscapes. Am Nat. 2015;85:212–27. https://doi.org/10.1086/679439.
Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–4. https://doi.org/10.1126/science.1165826.
Taylor MA, Cooper MD, Schmitt J. Phenological and fitness responses to climate warming depend upon genotype and competitive neighbourhood in Arabidopsis thaliana. Funct Ecol. 2019;33:308–22. https://doi.org/10.1111/1365-2435.13262.
Scheepens JF, Deng Y, Bossdorf O. Phenotypic plasticity in response to temperature fluctuations is genetically variable, and relates to climatic variability of origin, in Arabidopsis thaliana. AoB Plants. 2018;10:ply043. https://doi.org/10.1093/aobpla/ply043.
Lee U, Mortola EN, Kim EJ, Long M. Evolution and maintenance of phenotypic plasticity. BioSystems. 2022;222:104791. https://doi.org/10.1016/j.biosystems.2022.104791.
Murren CJ. The integrated phenotype. Integr Comp Biol. 2012;52:64–76. https://doi.org/10.1093/icb/ics043.
Huang X, Schmitt J, Dorn L, Griffith C, Effgen S, Takao S, et al. The earliest stages of adaptation in an experimental plant population: strong selection on QTLS for seed dormancy. Mol Ecol. 2010;19:1335–51. https://doi.org/10.1111/j.1365-294x.2010.04557.x.
Chiang GCK, Bartsch M, Barua D, Nakabayashi K, Debieu M, Kronholm I, et al. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol. 2011;20:3336–49. https://doi.org/10.1111/j.1365-294x.2011.05181.x.
Groot MP, Kubisch A, Ouborg NJ, Pagel J, Schmid KJ, Vergeer P, et al. Transgenerational effects of mild heat in Arabidopsis thaliana show strong genotype specificity that is explained by climate at origin. New Phytol. 2017;215:1221–34. https://doi.org/10.1111/nph.14642.
Zhang YY, Fischer M, Colot V, Bossdorf O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013;197:314–22. https://doi.org/10.1111/nph.12010.
Puy J, Carmona CP, Dvořáková H, Latzel V, de Bello F. Diversity of parental environments increases phenotypic variation in Arabidopsis populations more than genetic diversity but similarly affects productivity. Ann Bot. 2021;127:425–36. https://doi.org/10.1093/aob/mcaa100.
Picó FX. Variation and plasticity in life-history traits and fitness of wild Arabidopsis thaliana populations are not related to their genotypic and ecological diversity . 2024. Dryad. https://doi.org/10.5061/dryad.98sf7m0qp.
Acknowledgements
We thank David Ragel, Mercedes Ramiro, Bernardo Toledo, Marisol Muñoz, and the staff of the El Castillejo Botanical Garden for field, laboratory and/or greenhouse assistance. We also thank Maddie James and one anonymous reviewer for their comments on an earlier version of this manuscript. The Laboratorio de Ecología Molecular (LEM) from Estación Biológica de Doñana and the Doñana’s Singular Scientific and Technical Infrastructure (ICTS-RBD) provided logistical support. We thank the administration of the Sierra de Grazalema for permission to work at the El Castillejo Botanical Garden.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This research was funded by grants CGL2016-77720-P and PID2019-104135GB-I00 to FXP, and grant PID2022-136893NB-I00 to CA-B, from the Agencia Estatal de Investigación of Spain (AEI/10.13039/501100011033) and the Fondo Europeo de Desarrollo Regional (FEDER, UE).
Author information
Authors and Affiliations
Contributions
F.X.P. and C.A.B. conceived the idea. B.M.V., C.A.B., R.G., A.R.C. and F.X.P. generated materials and data. A.M.M., M.N., R.T.P., J.C.O. and C.A.B. generated or analyzed whole-genome sequences. R.dlM., A.M. and F.X.P. analyzed the data. F.X.P. wrote the first version of the manuscript and all authors contributed to it.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The species sampled is not endangered or protected.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
de la Mata, R., Mollá-Morales, A., Méndez-Vigo, B. et al. Variation and plasticity in life-history traits and fitness of wild Arabidopsis thaliana populations are not related to their genotypic and ecological diversity. BMC Ecol Evo 24, 56 (2024). https://doi.org/10.1186/s12862-024-02246-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-024-02246-x