Abstract
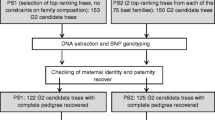
Polycross mating systems are widely used in forest tree breeding for genetic testing. Backward selection based on polycross testing assumes equal male reproductive success and true half-sib progeny. The main objectives of this study were, firstly, to investigate the departure from these assumptions in a maritime pine polycross trial and, secondly, to evaluate the consequences for heritability and breeding values estimations. A total of 984 offspring from 98 half-sib families was genotyped with single nucleotide polymorphism markers to recover the full pedigree. Paternity was assigned successfully for 89 % of the offspring at a 99 % confidence level. We thus concluded there was an 11 % pollen contamination rate, assuming contamination when no genotype from the polymix composition could be identified as a father. The paternal contribution to the offspring varied among the males, but the departure from half-sib assumption was moderate since the average genetic correlation within the family was 0.26. Heritability and breeding values for girth at breast height and stem sweep were estimated using individual-tree mixed models with either partial or full pedigree information. The results highlighted a minor bias in heritability estimation due to unknown paternity, as well as a high correlation for estimated breeding values between the partial and full pedigree models, suggesting that the genetic merit of the parental generation for backward selection was adequately predicted using the partial pedigree model. Finally, pedigree recovery was also discussed in a perspective of forward selection.




Similar content being viewed by others
References
Adams WT, Neale DB, Loopstra CA (1988) Verifying controlled crosses in conifer tree-improvement programs. Silvae Genet 37:147–152
Aronen T, Nikkanen T, Harju A, Tiimonen H, Haggman H (2002) Pollen competition and seed-siring success in Picea abies. Theor Appl Genet 104:638–642. doi:10.1007/s00122-001-0789-9
Burdon RD, Shelbourne CJA (1971) Breeding populations for recurrent selection: conflicts and possible solutions. N Z J For Sci 1:174
Canales J, Bautista R, Label P, Gomez-Maldonado J, Lesur I, Fernandez-Pozo N, Rueda-Lopez M, Guerrero-Fernandez D, Castro-Rodriguez V, Benzekri H, Canas RA, Guevara MA, Rodrigues A, Seoane P, Teyssier C, Morel A, Ehrenmann F, Le Provost G, Lalanne C, Noirot C, Klopp C, Reymond I, Garcia-Gutierrez A, Trontin JF, Lelu-Walter MA, Miguel C, Cervera MT, Canton FR, Plomion C, Harvengt L, Avila C, Gonzalo Claros M, Canovas FM (2014) De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology. Plant Biotechnol J 12:286–299. doi:10.1111/pbi.12136
Chancerel E, Lamy JB, Lesur I, Noirot C, Klopp C, Ehrenmann F, Boury C, Le Provost G, Label P, Lalanne C, Leger V, Salin F, Gion JM, Plomion C (2013) High-density linkage mapping in a pine tree reveals a genomic region associated with inbreeding depression and provides clues to the extent and distribution of meiotic recombination. BMC Biol 11:19. doi:10.1186/1741-7007-11-50
Doerksen TK, Herbinger CM (2008) Male reproductive success and pedigree error in red spruce open-pollinated and polycross mating systems. Can J For Res-Rev Can Rech For 38:1742–1749. doi:10.1139/x08-025
Doerksen TK, Herbinger CM (2010) Impact of reconstructed pedigrees on progeny—test breeding values in red spruce. Tree Genet Genomes 6:591–600. doi:10.1007/s11295-010-0274-1
Durel CE, Bertin P, Kremer A (1996) Relationship between inbreeding depression and inbreeding coefficient in maritime pine (Pinus pinaster). Theor Appl Genet 92:347–356
El-Kassaby YA, Cappa EP, Liewlaksaneeyanawin C, Klapste J, Lstiburek M (2011) Breeding without breeding: is a complete pedigree necessary for efficient breeding? PLoS One 6:11. doi:10.1371/journal.pone.0025737
Funda T, Liewlaksaneeyanawin C, El-Kassaby YA (2014) Determination of paternal and maternal parentage in lodgepole pine seed: full versus partial pedigree reconstruction. Can J For Res-Rev Can Rech For 44:1122–1127. doi:10.1139/cjfr-2014-0145
Gaspar MJ, de-Lucas A, Alia R, Paiva JAP, Hidalgo E, Louzada J, Almeida H, Gonzalez-Martinez SC (2009) Use of molecular markers for estimating breeding parameters: a case study in a Pinus pinaster Ait. progeny trial. Tree Genet Genomes 5:609–616. doi:10.1007/s11295-009-0213-1
Gauzere J, Oddou-Muratorio S, Pichot C, Lefevre F, Klein E (2013) Biases in quantitative genetic analyses using open-pollinated progeny tests from natural tree populations. Acta Bot Gall 160:225–236. doi:10.1080/12538078.2013.822827
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml user fuide release 3.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK
Glaubitz JC, Rhodes OE, Dewoody JA (2003) Prospects for inferring pairwise relationships with single nucleotide polymorphisms. Mol Ecol 12:1039–1047. doi:10.1046/j.1365-294X.2003.01790.x
Gonzalez-Martinez SC, Gerber S, Cervera MT, Martinez-Zapater JM, Alia R, Gil L (2003) Selfing and sibship structure in a two-cohort stand of maritime pine (Pinus pinaster Ait.) using nuclear SSR markers. Ann For Sci 60:115–121. doi:10.1051/forest:2003003
Grattapaglia D, Diener PSD, dos Santos GA (2014) Performance of microsatellites for parentage assignment following mass controlled pollination in a clonal seed orchard of loblolly pine (Pinus taeda L.). Tree Genet Genomes 10:1631–1643. doi:10.1007/s11295-014-0784-3
Hallingback HR, Jansson G (2013) Genetic information from progeny trials: a comparison between progenies generated by open pollination and by controlled crosses. Tree Genet Genomes 9:731–740. doi:10.1007/s11295-012-0588-2
Hansen OK, McKinney LV (2010) Establishment of a quasi-field trial in Abies nordmanniana—test of a new approach to forest tree breeding. Tree Genet Genomes 6:345–355. doi:10.1007/s11295-009-0253-6
Hansen OK, Nielsen UB (2010) Microsatellites used to establish full pedigree in a half-sib trial and correlation between number of male strobili and paternal success. Ann For Sci 67:10. doi:10.1051/forest/2010028
Illy G (1966) Recherches sur l’amélioration génétique du pin maritime. Ann Sci Forest 23:765–948. doi:10.1051/forest/19660401
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. doi:10.1111/j.1365-294X.2007.03089.x
Kimura M, Crow JF (1963) The measurement of effective population number. Evolution 17:279–288
Klapste J, Lstiburek M, El-Kassaby YA (2014) Estimates of genetic parameters and breeding values from western larch open-pollinated families using marker-based relationship. Tree Genet Genomes 10:241–249. doi:10.1007/s11295-013-0673-1
Korecky J, Klapste J, Lstiburek M, Kobliha J, Nelson CD, El-Kassaby YA (2013) Comparison of genetic parameters from marker-based relationship, sibship, and combined models in Scots pine multi-site open-pollinated tests. Tree Genet Genomes 9:1227–1235. doi:10.1007/s11295-013-0630-z
Kumar S, Gerber S, Richardson TE, Gea L (2007) Testing for unequal paternal contributions using nuclear and chloroplast SSR markers in polycross families of radiata pine. Tree Genet Genomes 3:207–214. doi:10.1007/s11295-006-0056-y
Kumar S, Richardson TE (2005) Inferring relatedness and heritability using molecular markers in radiata pine. Mol Breed 15:55–64. doi:10.1007/s11032-004-2059-4
Lai BS, Funda T, Liewlaksaneeyanawin C, Klapste J, Van Niejenhuis A, Cook C, Stoehr MU, Woods J, El-Kassaby YA (2010) Pollination dynamics in a Douglas-fir seed orchard as revealed by pedigree reconstruction. Ann For Sci 67:8. doi:10.1051/forest/2010044
Lambeth C, Lee BC, O’Malley D, Wheeler N (2001) Polymix breeding with parental analysis of progeny: an alternative to full-sib breeding and testing. Theor Appl Genet 103:930–943. doi:10.1007/s001220100627
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Inc., Sunderland, MA, USA, 980 p
Mangin B, Siberchicot A, Nicolas S, Doligez A, This P, Cierco-Ayrolles C (2012) Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 108:285–291. doi:10.1038/hdy.2011.73
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655. doi:10.1046/j.1365-294x.1998.00374.x
Moriguchi Y, Ishiduka D, Kaneko T, Itoo S, Taira H, Tsumura Y (2009) The contribution of pollen germination rates to uneven paternity among polycrosses of Cryptomeria japonica. Silvae Genet 58:139–144
Nakamura RR, Wheeler NC (1992) Pollen competition and paternal success in Douglas-fir. Evolution 46:846–851. doi:10.2307/2409655
Parantainen A, Pasonen HL (2004) Pollen-pollen interactions in Pinus sylvestris. Scand J Forest Res 19:199–205. doi:10.1080/02827580410029336
Pasonen HL, Pulkkinen P, Kapyla M, Blom A (1999) Pollen-tube growth rate and seed-siring success among Betula pendula clones. New Phytol 143:243–251. doi:10.1046/j.1469-8137.1999.00451.x
Plomion C, Chancerel E, Endelman J, Lamy JB, Mandrou E, Lesur I, Ehrenmann F, Isik F, Bink M, van Heerwaarden J, Bouffier L (2014) Genome-wide distribution of genetic diversity and linkage disequilibrium in a mass-selected population of maritime pine. BMC Genomics 15:16. doi:10.1186/1471-2164-15-171
RCoreTeam (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
Snow AA, Spira TP (1996) Pollen-tube competition and male fitness in Hibiscus moscheutos. Evolution 50:1866–1870. doi:10.2307/2410744
Squillace AE (1974) Average genetic correlations among offspring from open-pollinated forest trees. Silvae Genetica 23:149–156
Wheeler N, Payne P, Hipkins V, Saich R, Kenny S, Tuskan G (2006) Polymix breeding with paternity analysis in Populus: a test for differential reproductive success (DRS) among pollen donors. Tree Genet Genomes 2:53–60. doi:10.1007/s11295-005-0033-x
White T (1987) A conceptual framework for tree improvement programs. New For 1:325–342. doi:10.1007/BF00031742
Wiselogel AE, Vanbuijtenen JP (1988) Probability of equal mating in polymix pollinations of loblolly pine (Pinus taeda L.). Silvae Genet 37:184–187
Zhao P, Zhang SX, Woeste K (2013) Genotypic data changes family rank for growth and quality traits in a black walnut (Juglans nigra L.) progeny test. New For 44:357–368. doi:10.1007/s11056-012-9343-7
Zobel BJ, Talbert JT (1984). Applied forest tree improvement (p. 528). New York: Wiley.
Acknowledgments
This study was funded by INRA (EFPA division projet innovant), the European-Union (ProCoGen project: n° 289841), and Conseil Regional d’Aquitaine (IMAF project co-funded by FCBA: n° 120009468–052). Marjorie Vidal received a CIFRE PhD fellowship (public/private research partnerships between FCBA and the French Ministry of Higher Education and Research). This study would not have been possible without the support of the maritime Pine Breeding Cooperative (GIS Pin Maritime du Futur). We gratefully acknowledge all its members. The authors also thank the INRA Experimental Unit (UE0570) for field measurements, Sophie Gerber (INRA) for helpful discussions on paternity recovery, Adline Delcamp (INRA) for the genotyping stage, Jean-Mathieu De Boisseson (FCBA) for needle sampling, Maël Ruby for his preliminary study on paternity recovery, and Fikret Isik (NCSU, USA) for his advice in shaping the content of the paper and his help in ASReml analysis.
The genotyping was performed at the Genomic Facility of Bordeaux (grants from the Conseil Regional d’Aquitaine: n°20030304002FA and 20040305003FA, the European Union: FEDER n°2003227 and ANR: n°ANR-10-EQPX-16 Xyloforest).
Data archiving statement
All SNPs used in this study were deposited in dbSNP (NCBI database), available on http://www.ncbi.nlm.nih.gov/.
The supplemental data “Supplemental_data_SNP_description.txt” contains the dbSNP accession ID for each SNP (#ss) and some useful information (SNP ID, set identity, contig name, linkage group LG, position on the LG, SNP type, and sequence surrounding the SNP).
The genotypes of G2 trees and their potential parents (G1 trees) are available in the supplemental data file “Subset_A_genotypes_56SNPs.csv” for Subset A and “Subset_B_genotypes_63SNPs.csv” for subset B. The first column contains the individual IDs, and the first line contains the SNP IDs. Blank cells are missing data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: J. Beaulieu
This article is part of the Topical Collection on Breeding
Rights and permissions
About this article
Cite this article
Vidal, M., Plomion, C., Harvengt, L. et al. Paternity recovery in two maritime pine polycross mating designs and consequences for breeding. Tree Genetics & Genomes 11, 105 (2015). https://doi.org/10.1007/s11295-015-0932-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0932-4