Abstract
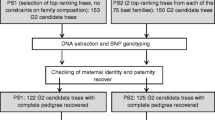
Pedigrees reconstructed through DNA marker assigned paternities in polymix (PMX) and open pollinated (OP) progeny tests were analyzed using mixed models to test the effect of unequal male reproductive success and pedigree errors on quantitative genetic parameters. The reconstructed pedigree increased heritabilities in the larger PMX test. Increased heritability resulted from adding the paternities to the pedigree per se, not by correcting the male reproductive bias by specifying the exact pedigree. Removing hypothesized pedigree errors had no effect on quantitative parameters, either because the magnitude of the errors was too small (PMX) or the progeny test was too small to detect variance components reliably (OP). Although there was no advantage in backwards selection, the increased additive variance, heritabilities and accuracy of progeny with assigned paternities in the pedigree, should permit forward selection of offspring with greater genetic gain and complete control of coancestry for future breeding decisions. Some possible breeding population structures with the new genetic information are discussed.


Similar content being viewed by others
References
Banos G, Wiggans GR, Powell RL (2001) Impact of paternity errors in cow identification on genetic evaluations and international comparisons. J Dairy Sci 84(11):2523–2529
Bernardo R (1993) Estimation of coefficient of coancestry using molecular markers in maize. Theor Appl Genet 85(8):1055–1062
Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34(1):20–25
Brisbane JR, Gibson JP (1995). Balancing selection response and rate of inbreeding by including genetic relationships in selection decisions. Theor Appl Genet 91(3):421–431
Bromley CM, Van Vleck LD, Johnson BE, Smith OS (2000) Estimation of genetic variance in corn from F1 performance with and without pedigree relationships among inbred lines. Crop Sci 40(3):651–655
Burczyk J, Adams WT, Moran GF, Griffin AR (2002) Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighbourhood model. Mol Ecol 11(11):2379–2391
Burdon RD, Kumar S (2004) Forwards versus backwards selection: trade-offs between expected genetic gain and risk avoidance. N Z J For Sci 34(1):3–21
Cotterill PP (1986) Genetic gains expected from alternative breeding strategies including simple low cost options. Silvae Genet 35(5–6):212–223
Cotterill PP (1989). The nucleus breeding system. In: Proceedings of the twentieth southern forest tree improvement conference. Charleston, SC, pp 36–42
de Souza VA, Byrne DH, Taylor JF (2000) Predicted breeding values for nine plant and fruit characteristics of 28 peach genotypes. J Am Soc Hortic Sci 125(4):460–465
Dodds KG, Tate ML, Sise JA (2005) Genetic evaluation using parentage information from genetic markers. J Anim Sci 83(10):2271–2279
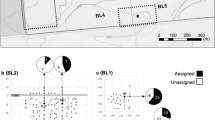
Doerksen TK, Herbinger CM (2008) Male reproductive success and pedigree error in red spruce open-pollinated and polycross mating systems. Can J For Res 38(7):1742–1749
Durel CE, Laurens F, Fouillet A, Lespinasse Y (1998) Utilization of pedigree information to estimate genetic parameters from large unbalanced data sets in apple. Theor Appl Genet 96(8):1077–1085
El-Kassaby YA, Lstibůrek M, Liewlaksaneeyanawin C, Slavov GT, Howe GT (2006) Breeding without breeding: approach, example, and proof of concept. In: Isik F, (ed) Proceedings of the IUFRO division 2 joint conference: low input breeding and conservation of Forest genetic resources. Antalya, Turkey, pp. 43–54. (verified: 03/Feb/2007)
Ericsson T (1999) The effect of pedigree error by misidentification of individual trees on genetic evaluation of a full-sib experiment. Silvae Genet 48:239–242
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Longman, Harlow, England
Fowler DP (1986) Strategies for the genetic improvement of important tree species in the Maritimes. Information Report M-X-156, Canadian Forestry Service
Frensham AB, Barr AR, Cullis BR, Pelham SD (1998) A mixed model analysis of 10 years of oat evaluation data: use of agronomic information to explain genotype by environment interaction. Euphytica 99(1):43–56
Hadfield JD, Richardson DS, Burke T (2006) Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol Ecol 15(12):3715–3730
Henderson CR (1988) Use of an average numerator relationship matrix for multiple-sire joining. J Anim Sci 66(7):1614–1621
Hodge GR, Volker PW, Potts BM, Owen JV (1996) A comparison of genetic information from open-pollinated and control-pollinated progeny tests in two eucalypt species. Theor Appl Genet 92(1):53–63
Israel C, Weller JI (2000) Effect of misidentification on genetic gain and estimation of breeding value in dairy cattle populations. J Dairy Sci 83(1):181–187
Jackson N (1983) Effect of ignoring full sib relationships when making half sib estimates of heritability. Theor Appl Genet 65(1):61–66
James JW (1977) Open nucleus breeding systems. Anim Prod 24(3):287–305
John JA, Williams ER (1998) t-latinized designs. Aust N Z J Stat 40(1):111–118
Joyce D, Ford R, Fu YB (2002) Spatial patterns of tree height variations in a black spruce farm-field progeny test and neighbors-adjusted estimations of genetic parameters. Silvae Genet 51(1):13–18
Klein JI (1995). Multiple-trait combined selection in jack pine family-test plantations using best linear prediction. Silvae Genet 44(5–6):362–375
Kumar S (2006) Correlation between clonal means and open-pollinated seedling progeny means and its implications for radiata pine breeding strategy. Can J For Res 36(8):1968–1975
Lambeth C, Lee BC, O’Malley D, Wheeler N (2001) Polymix breeding with parental analysis of progeny: an alternative to full-sib breeding and testing. Theor Appl Genet 103(6):930–943
Lindgren D (1975) Use of selfed material in forest tree improvement. Research Notes 15, The Royal College of Forestry, Department of Forest Genetics, Stockholm, Sweden
Lindgren D, Mullin TJ (1997) Balancing gain and relatedness in selection. Silvae Genet 46(2–3):124–129
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates Inc., Sunderland, MA, USA
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7(5):639–655
Meyer K (2006) WOMBAT: a program for mixed model analyses by restricted maximum likelihood. Animal Genetics and Breeding Unit, Armidale, Australia. User notes, 58pp
Milner JM, Brotherstone S, Pemberton JM, Albon SD (2000) Estimating variance components and heritabilities in the wild: a case study using the ‘animal model’ approach. J Evol Biol 13(5):804–813
Mrode R (2006) Linear models for the prediction of animal breeding values. CABI, Wallingford, UK
Piepho HP, Möhring J, Melchinger AE, Büchse A (2008) BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161(1–2):209–228
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Ruotsalainen S, Lindgren D (1998) Predicting genetic gain of backward and forward selection in forest tree breeding. Silvae Genet 47(1):42–50
Shaw DV, Hood JV (1985) Maximizing gain per effort by using clonal replicates in genetic tests. Theor Appl Genet 71(3):392–399
Sorensen FC, White TL (1988) Effect of natural inbreeding on variance structure in tests of wind pollination Douglas-fir progenies. For Sci 34(1):102–118
Squillace AE (1974) Average genetic correlations among offspring from open-pollinated forest trees. Silvae Genet 23(5):149–156
Van Vleck LD (1970) Misidentification in estimating the paternal sib correlation. J Dairy Sci 53(10):1469
Visscher PM, Woolliams JA, Smith D, Williams JL (2002) Estimation of pedigree errors in the UK dairy population using microsatellite markers and the impact on selection. J Dairy Sci 85(9):2368–2375
Wheeler N, Payne P, Hipkins V, Saich R, Kenny S, Tuskan G (2006) Polymix breeding with paternity analysis in Populus: a test for differential reproductive success (DRS) among pollen donors. Tree Genetics & Genomes 2(1):53–60
White TL, Hodge GR (1988) Best linear prediction of breeding values in a forest tree improvement program. Theor Appl Genet 76(5):719–727
Williams CG, Savolainen, O. (1996) Inbreeding depression in conifers: implications for breeding strategy. For Sci 42(1):102–117
Wilson AJ, Mcdonald G, Moghadam HK, Herbinger CM, Ferguson MM (2003) Marker-assisted estimation of quantitative genetic parameters in rainbow trout, Oncorhynchus mykiss. Genetics Research 81(02):145–156
Wright AJ (1980) The expected efficiencies of half-sib, testcross and S1 progeny testing methods in single population improvement. Heredity 45(3):361–376
Yanchuk AD (2001) A quantitative framework for breeding and conservation of forest tree genetic resources in British Columbia. Can J For Res 31:566–576
Acknowledgements
We thank two anonymous reviewers for comments that improved the article. We thank Svetlana Shkuratova, Kyle Gardner and Sarah Reid at Dalhousie University; Howard Frame and Dave Steeves at the Nova Scotia Department of Natural Resources (Truro); and Rick Allen, Derek Geldart and Kari Easthouse at NewPage Corp. (formerly Stora-Enso, Port Hawkesbury). Funding was provided by an Atlantic Innovation Fund (ACOA) grant to the second author (CMH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Rights and permissions
About this article
Cite this article
Doerksen, T.K., Herbinger, C.M. Impact of reconstructed pedigrees on progeny-test breeding values in red spruce. Tree Genetics & Genomes 6, 591–600 (2010). https://doi.org/10.1007/s11295-010-0274-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0274-1