Abstract
Cuticular waxes were extracted from the leaves of a coconut mapping population generated by the controlled cross of an East African Tall and a Rennell Island Tall genotype for the construction of molecular linkage maps. The wax composition was analyzed by capillary gas chromatography/mass spectrometry, and for eight of the wax compounds, their absolute and relative amounts were determined. As reported previously for a different coconut ecotype (Malayan Yellow Dwarf), lupeol methyl ether, isoskimmiwallin, and skimmiwallin were identified as the major components of coconut cuticular wax. The additional compounds were characterized as 3-β-methoxy lupane (lupane methyl ether), lupeol and the acetic acid esters of lupeol, skimmiwallinol, and isoskimmiwallinol, respectively. Minor, nonidentified compounds amounted to some 5% of total wax content and included triterpenoids, sterols, primary alcohols, and fatty acids. The variation detected for parents and progeny with respect to the wax components allowed quantitative trait locus (QTL) analyses for their biosynthetic pathways. A total of 46 QTLs could be mapped onto the coconut linkage map which was extended by amplified fragment length polymorphism and single sequence repeat markers into a high density map with more than 1,000 mapped DNA markers. Several colocated QTLs for different traits were detected reflecting the observed correlations among characters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary surfaces of vascular plants are covered by the cuticle, a continuous extracellular membrane that represents the interface of the plant to its aerial environment. The cuticle is mainly composed of a matrix of polymeric oxygenated C16 and C18 fatty acid derivatives cross-linked via ester bonds (for reviews see Jeffrey 1996; Post-Beittenmiller 1996; Kolattukudy 1998; Sommerville et al. 2000). Into the polymer matrix and at its outer surface mostly monomeric lipids are embedded as intracuticular waxes or superimposed as epicuticular waxes, respectively. The components most commonly found in the cuticular wax of higher plants include very long chain n-alkanes, ketones, alcohols, aldehydes, esters, and terpenoids. For example, in Arabidopsis, wax components mostly consist of aliphatic chains 20–30 carbons in length as well as of primary and secondary alcohols, aldehydes, and ketones (Lemieux et al. 1994).
Waxes are involved in the plant’s defense against abiotic stress: The strongly hydrophobic waxes play an important role in water retention by limiting nonstomatal water loss (Kerstiens 1996; Riederer and Schreiber 2001; Jenks 2002). Secondly, the crystalline structure of surface wax is involved in light reflection as obvious from glossy mutants, and changes in crystal structure with concomitant changes in light reflection may help the plant in protection from damaging UV light or in the adaptation to different light intensities. Waxes also form part of the preformed plant defense system against biotic stresses such as insects, bacteria, and fungi (Gülz et al. 1991; Yoon et al. 1998; Marcell and Beattie 2002). In particular, their chemical makeup and abundance are known to affect resistance to insects (Kolattukudy 1987).
Such protective role of epicuticular wax has also been reported in coconut (Cocos nucifera) by Kurup et al. (1993) who observed a clear indication of a negative relationship between the epicuticular wax content of coconut leaves and transpiration rate. In general, the epicuticular wax content was higher among drought tolerant than among susceptible genotypes. Independently of the genotype, however, the content increased sharply during periods of water stress and declined in poststress periods. This peak coincided with high light intensity, high temperature, and low relative humidity. The composition of cuticular wax in coconut has been studied to an only limited extent. It was recently reported for two single coconut genotypes that lupeol methyl ether, isoskimmiwallin, and the skimmiwallin are the major components of coconut epicuticular wax (Escalante et al. 2002). In this publication, quantitative trait locus (QTL) analyses for wax content and wax components were performed on a mapping population derived from a cross between the coconut genotypes East African Tall (EAT) and Rennell Island Tall (RIT). Further five wax components could be identified in cuticular wax of C. nucifera and the corresponding QTLs were mapped onto an integrated molecular linkage map.
Material and methods
Plant material
The coconut mapping population originated from controlled pollinations of an East African Tall palm (EAT1011) with pollen from a Rennell Island Tall palm (RIT0710). Both parental genotypes were heterozygous. A progeny of 94 genotypes was established and field-planted in 1994 at Chambezi Research Station (Tanzania) under the auspices of the Mikocheni Agricultural Research Institute (MARI). The station is located near the coast around 80 m above sea level and has sandy soils. Average annual rainfall is around 1,060 mm with a marked rain season from March to May (60% of annual precipitations). Average monthly minimum temperatures vary between 19° and 25°C and maximum temperatures between 28°C and 31°C.
Wax analysis
Wax contents
Six leaflets were collected from the 10th frond of each progeny genotype, carefully cleaned, and cut into pieces of approximately 25 cm length, resulting in 12 leaf samples per genotype. For wax extraction, their midribs were removed, and for each sample, the cuticular wax was extracted into 80 ml of hexane in a plastic beaker during 2 min at ambient temperature. After recovering the leaf samples, the organic solution was filtered and transferred to new plastic cups. Wax samples were dried overnight under the hood followed by heating at 50°C until constant weight. The wax amount was determined by weight determination. The recovered leaf samples were dried and placed on a scanner for leaf area determination using fixed area standards and in-house developed software.
Wax component analyses
Dried wax samples of each progeny genotype were dissolved in 1 ml of chloroform each. A defined fraction of these solutions was transferred to sample vials and n-tetracosane (5 μg) was added as internal standard. Prior to analysis, chloroform was evaporated from all samples under a gentle stream of N2 while heating the sample vials to 50°C. Then all samples were treated with bis-N,N-(trimethylsilyl)trifluoroacetamide in pyridine for 30 min at 70°C to transform all hydroxyl-containing compounds to the corresponding trimethylsilyl derivatives. Afterwards, the final volume (140 μl) was adjusted with chloroform.
The qualitative composition was studied by capillary gas chromatography (GC; 6890N; Agilent Technologies, Palo Alto, CA, USA; 30 m DB-1, 0.32 mm i.d.; df = 0.1 μm; J&W Scientific, Folsom, CA, USA) and mass selective detection (5973N; Agilent; 70 eV, m/z 50–750). The inlet pressure of the carrier gas (Helium 5.0; Messer Griesheim, Krefeld, Germany) at the time of injection was at 50 kPa and maintained for 5 min, before increasing the pressure at a rate of 3 kPa min−1 to 150 kPa. This pressure was maintained up to the end of the temperature program. GC was carried out with temperature-programmed injection (on-column) at 50°C, oven 2 min at 50°C, raised by 40°C min−1 to 200°C, held for 2 min at 200°C, raised by 3°C min−1 to 320°C, and held for 30 min at 320°C. The quantitative composition of the mixtures was studied by capillary GC (5890 II; Hewlett Packard, Avondale, PA, USA; column as above) and flame ionization detection under the same GC conditions as above, but with hydrogen (Hydrogen generator; Whatman, Haverhill, MA, USA) as the carrier gas. Single compounds were quantified against the internal standard by manually integrating peak areas.
Each chromatogram was intensively examined for contaminations with plasticizers resulting from the extraction procedure in plastic beakers. Except for traces of one phthalate derivative, no other contamination could be detected. Analyses of blank samples (with and without palm waxes) obtained by using only glass materials revealed this plasticizer to be inherent to the used GC-Systems.
DNA extraction
Total DNA was extracted from leaf material using a modified cetyltrimethylammonium bromide (CTAB) method of Doyle and Doyle (1990). The basic CTAB buffer was supplemented with 2% polyvinyl pyrrolidone 40, 25 mM sodium ascorbate, 25 mM sodium bisulfite, and 10 mM diethyldithiocarbamate (Ramírez et al. 2004). Five grams of leaf material were frozen in liquid nitrogen and converted to a fine powder by blending in a coffee mill or technical blender. After the addition to 25 ml of CTAB buffer prewarmed at 65°C, the suspension was briefly shaken and extracted twice with equal volumes of chloroform and subsequent phase separation by centrifugation. Nucleic acids were precipitated from the aqueous phase by the addition of 0.8 volumes of isopropanol, collected by centrifugation, and dissolved in TE buffer (10 mM TRIS-HCl, pH 8, 0.1 mM EDTA) for RNAse treatment, phenol extraction, and ethanol precipitation as described previously (Rohde 1996). Precipitated DNA was dissolved in TE, and its integrity and concentration were determined by electrophoresis in 0.7% agarose gels and comparison to a 1 kb DNA ladder standard (GIBCO-BRL, Netherlands).
DNA marker analysis
Amplified fragment length polymorphism (AFLP) analysis was performed according to Vos et al. (1995). Genomic DNA was digested using EcoRI and MseI, and EcoRI and MseI adapters were ligated to the restricted DNA. Preamplification was performed with primers containing one selective nucleotide and specific amplification with primers containing three selective nucleotides. Primers were either labeled with the fluorescent infrared dyes IRD800 or IRD700 or alternatively 33P-labeled AFLP primers were used. Amplification products were separated on 4% polyacrylamide gels under denaturing conditions. Fluorescent AFLP fragments were detected on a LI-COR 4200-S1 DNA sequencer (LI-COR, Lincoln, NE, USA). Analysis was performed according to the manufacturer’s instructions. Radioactively labeled fragments were fixed on the gel in 10% acetic acid, washed with water, dried, and exposed to X-rays films at room temperature for 1 to 3 days. Single sequence repeat (SSR) analysis was performed as described by Lebrun et al. (2001). Amplified alleles were revealed by silver staining or using fluorescence-labeled primers with the LI-COR system. Both coconut and heterologous SSR markers from oil palm were used (Billotte et al. 2005). Finally, COS clones were anchored to mapped AFLP markers using multidimensional pools of COS clones as described by Sniady et al. (2003).
Computational methods and statistics
Data analysis and linkage mapping
For data processing, polymorphic DNA fragments were scored for presence or absence in parents and progeny genotypes. Map construction was performed as described in detail by Ritter et al. (1990) and Ritter and Salamini (1996). This procedure involves linkage analysis between marker fragments, estimation of recombination frequencies, and determination of linear order between linked loci applying multipoint linkage analysis and the expectation–maximization algorithm for handling missing data. Data were processed for linkage mapping with the MAPRF program (Ritter and Salamini 1996). Initially, independent parental linkage maps were constructed considering only segregating parent-specific fragments. Firstly, linkage subgroups were established composed of closely linked fragments with a minimum logarithm of the odds (LOD) threshold of 5.0 with at least one other fragment of the subgroup. Within each linkage subgroup, fragments were ordered by minimizing the sum of LOD scores for alternative orders. Afterwards, larger contiguous linkage groups (LGs) were established by connecting distal markers of appropriate subgroups based on maximum LOD values/minimum recombination frequencies between them. Finally, fragments common to both parents were integrated into linkage groups of both parental linkage maps as anchor points. These fragments—together with SSR markers—were used to combine the parental linkage maps as described by Ritter et al. (1990).
QTL analyses
QTLs were mapped using the least square interval mapping method developed for backcross progenies (Knapp et al. 1990; Knapp and Bridges 1990) in a similar way as described by Baudouin et al. (2006) for the common parent of a halfsib mapping population. In our study, all marker intervals of both individual parental linkage maps were used to identify genomic regions controlling the traits mentioned above. The SAS procedure PROC NLIN (SAS Institute Inc 1989) was used for computational analysis. Multiple regression analysis on all QTL intervals of each trait was applied to calculate the percentage of total variance explained by the sum of the individual QTLs. SAS software was also used to calculate Pearson coefficients for the correlation analyses between traits.
Results
Qualitative and quantitative analyses of coconut cuticular wax
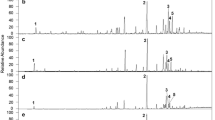
Total leaf cuticular waxes were extracted from the progeny genotypes of the mapping population by immersion of leaflets into n-hexane. These extracts were subjected to GC/mass spectrometry analyses as described in “Materials and methods”. In all extracts, eight compounds (I–VIII; Fig. 1) comprised approximately 95% of the cuticular waxes. The three main components (cf. Table 1) could be identified as lupeol methyl ether (LME; I), isoskimmiwallin (IS; V), and skimmiwallin (SW; VI) by comparing the corresponding mass spectra (Fig. 2) with those already described for a different coconut ecotype and other Palmae (Escalante et al. 2002; Garcia et al. 1995). The mass spectra of compounds VII and VIII (Fig. 2) strongly resembled those of IS and SW but showed a shift of [M]+ and a corresponding [M-15]+ to m/z 510 and m/z 495, respectively. Most recently, these compounds were identified as the acetylated derivatives of the parent metabolites isoskimmiwallinol and skimmiwallinol, namely isoskimmiwallinol acetate (ISA; VII) and skimmiwallinol acetate (SWA; VIII; Escalante et al. unpublished). In turn, the mass spectra of compounds II, III, and IV showed fragmentation patterns of lupane- and lupene-type triterpenoids (Fig. 2) that could be assigned to lupane methyl ether (MLU; II), lupeol (LUP; III; trimethylsilylated derivative), and its acetylated derivative lupeol acetate (LAC; IV) by comparison with spectra of authentic standards.
Absolute and relative variation of total amounts of wax and wax components were analyzed in the progeny genotypes (Table 1). Wax contents between progeny genotypes varied largely between 50.3 and 487.9 μg/dm2 with an average value of 195.1 μg/dm2 and a coefficient of variation (CV) of 55.2%.
Wax component analyses showed that lupeol methyl ether is the main wax component with an average 57.4% representation and a variation from 41% to 77%. Other major components are skimmiwallin (11.9%), isoskimmiwallin (8.6%), and lupeol acetate (5.5%). Other components such as lupeol, 3-β-methoxy lupane, isoskimmiwallinol acetate, and skimmiwallinol acetate are present in fractions between 1% and 3%. Triterpenoids and sterols represent 5% of the wax fraction, and other compounds (OC; primary alcohols and fatty acids) appear in negligible quantities (0.1%). The coefficients of variations for the main compound values varied between 46% and 92% and were generally much smaller for the relative values, except for MLU. Relative CVs were smaller for LME (17%) and lupeol (23%) and larger for the other specific compounds (between 39% and 52%). The CV of OC contents was extremely high with 260%, since these other substances were only found in certain genotypes.
Table 2 shows the results of the trait correlation analyses. Total wax amount and absolute wax component values were significantly positively correlated with each other, except for the contents of other compounds, which did not show any relationship with the other characters. This pattern changes drastically considering the relative wax composition. Wax contents was only significantly negatively correlated with the minor wax compound MLU. Lupeol methyl ether showed significantly negative correlations with all compounds of the skimmiwallin group. Skimmiwallin and its derivates presented positive correlations among most of them. Again, OC contents did not show any correlation with other compounds.
Establishment of a molecular linkage map
A total of 241 AFLP primer combinations were evaluated on parents and progeny genotypes of the mapping population EAT1011×RIT0710. Individual combinations generated between one and 20 segregating fragments and 1,503 polymorphic AFLP fragments were obtained in total. The EAT parent (879 fragments) was more polymorphic than the RIT parent (452 fragments). A total of 163 fragments were common to both parents. For nine fragments, the parental pattern was ambiguous and they were discarded. Furthermore, 64 coconut SSR primer combinations were analyzed in parents and progeny genotypes of the mapping population. Forty-eight of them produced segregating polymorphisms, while 16 were not polymorphic or generated unclear patterns. In addition, 22 heterologous SSR primers (originally developed for oil palm) were assayed with seven of them being functional and amplifying between one and four segregating alleles. Several coconut and one oil palm SSR generated two loci each.
Initially, individual parental linkage maps with 16 linkage groups each (in accordance with the 16 chromosomes of the haploid coconut genome) were obtained based on individual parent-specific fragments for the two parents EAT1011 and RIT0710 of the mapping population (results not shown). Markers common to both parents as well as the polymorphic, codominant SSR markers allowed the alignment of both parental linkage maps into an integrated map. Table 3 shows the characteristics of this map: A total of 704 markers were placed onto the integrated map resulting in a total map length of 2,739 cM. Individual linkage groups vary between 105 and 201 cM in length and contain between 25 and 61 markers each (average of 44 markers per LG). In addition, a total of 167 so-called RF0 markers (i.e., markers which are linked with a recombination frequency (RF) of zero to other mapped markers) were identified, but are not displayed for reasons of clarity. Furthermore, 93 so-called associated markers were determined. These markers did not fit precisely the existing framework maps (probably due to scoring errors), but showed reduced RF values (<10 cM) with other mapped markers and, therefore, were scored as “associated” markers. Thus, the integrated EAT/RIT map (Fig. 3) contains 1,026 DNA markers (including RF0 and associated fragments) with an average of 64 markers per linkage group.
Integrated linkage map of the cross EAT1011×RIT0710. Distances are given in cM (Kosambi units). QTLs are integrated into the map and shown in bold and underlined. Individual markers descending from RIT0710 are indicated in italics, markers common to both parents are underlined and markers from EAT1011 are indicated with normal font. SSR markers (with prefix ‘m’) and anchored COS clones are given in bold. For QTL names, see Table 1. For other QTL, marker names, and more details, see: http://www.neiker.net/link2palm
The mapped markers include 53 coconut SSR loci located on 14 linkage groups and eight oil palm SSR loci on six linkage groups, covering in total 15 of the 16 linkage group with SSR markers. Also 14 COS clones could be mapped to AFLP markers, assaying a total of 23 AFLP primer combinations on DNAs from the 30 pools of COS clones as described previously by Sniady et al. (2003). Furthermore, differences in the alleles of the desaturase gene (ACDP1; data not shown) revealed segregating polymorphism in the progeny and allowed to map this gene to linkage group 3 (ACPD1). Details on markers, parental linkage maps RF0 and associated markers can be found at http://www.neiker.net/Link2palm/DEFcoco.htm.
QTL analysis
The coefficients of variations and minimum and maximum values showed sufficient variation which allowed the detection of QTLs for all traits under study (Table 1). A total of 46 putative QTLs were identified for wax contents and wax component traits at a significance level of 5% and their number varied from one for ISA contents and eight for SWA contents. Total variance explained by the sum of all individual QTLs was relative low for ISA (10%) and TS contents (24%) due to the small number of QTLs, but ranged between 39 and 65% for all other characters under study. Therefore, markers linked to the detected QTLs for these traits could be used efficiently for marker-assisted selection.
Table 4 describes in detail the individual QTLs for each trait, which are also displayed on the linkage map (Fig. 3). QTLs were found on 14 of the 16 coconut linkage groups and no QTLs were present on LG1 and LG15. In contrast, eight QTLs are located on LG6 and LG10, respectively. Only ten of the 46 QTLs for wax composition descend from RIT. No QTL from RIT were detected for total wax amount but two of the three QTLs of the major wax compound LME originate from this parent. Individual QTLs explain between 5.9% and 32% of the total variation.
Four QTLs descending from the EAT parent were detected for wax contents and explain between 10.5% and 17.0% of the total variance. They are located on three different linkage groups (Table 4) and are always linked to several QTLs of single wax compounds. This is the case for qWXa on LG6 where wax amount is linked to QTLs for mayor components LME and LAC and on LG8 where qWXb is colocated with a QTL for SW contents. The two wax QTLs on LG10 are linked to IS/TS QTls and to SW/TS QTLs, respectively. In all cases, the relative effects of the QTLs are in the same direction (Table 4); i.e., with increasing wax contents, also the absolute contents of the different compounds increases. No QTL was detected near the desaturase gene ACPD1 which converts stearic acid into oleic acid by introducing a cis double bond at the C9 position of the C18 fatty acid.
In several other cases, QTLs for different wax compounds can be found closely linked or at identical genomic locations. For example, we find in several cases relationships between the acetylated and nonacetylated compounds such as the QTLs for SW and SWA contents on LG6, LG8, and LG10 and QTLs for LUP and LAC on LG4 and LG6. On LG4, we can observe colocation of QTLs for LUP and SWA and nearby a QTL for LAC. On LG11, qSWAh and qLACf are colocated and nearby, we can observe a QTL for ISA contents. This was the only case were an opposite effect was detected (i.e., ISA contents decreases for increasing SWA and LAC contents). On the other hand, the four QTLs for OC content and the only QTL for ISA are located at distinct genomic regions.
In some cases, SSR markers are located at QTLs or nearby. Examples are the microsatellite markers mCN125 and mCnE10 on LG5, mCN61 on LG6, and the oil palm SSR mEg3620 on LG8 which is colocated with qLUPc on LG9. In other cases (Fig. 3), QTLs can be found at the same position or close to mapped COS clones. This is the case for COS3 and qSWAb on LG5, or for COS2 and qLUPb on LG6.
Discussion
This work represents the first published attempt to identify QTLs for wax contents and its major components in coconut.
Composition of cuticular waxes
In most seed plant species, cuticular waxes comprise a complex mixture of mainly aliphatic compounds like n-alkanes, primary and secondary alcohols, fatty acids, and n-aldehydes (for reviews see Jetter et al. 2006; Walton 1990; Baker 1982). In many plant taxa, also cyclic compounds like triterpenoids, sterols, and phenols can be found to a limited extent while only in a few species triterpenoids represent the dominating compound class (e.g., Guhling et al. 2006; Markstädter et al. 2000; Hemmers and Gülz 1989).
To date, about 200 different basic chemical structures of triterpenoids from natural sources are described (Xu et al. 2004), with pentacyclic triterpenoids being the most common in plant cuticular waxes (Walton 1990). The majority in terms of structural diversity and relative contribution to wax composition is derived from lupane, oleanane, and ursane backbones with a hydroxyl function in the 3β-position (Jetter et al. 2006; Walton 1990). For example, lupeol was identified as the forming compound of epicuticular wax crystals in Ricinus communis (Guhling et al. 2006) while in Prunus laurocerasus, oleanoic and ursolic acid only accumulate in the intracuticular wax compartment (Jetter and Schäffer 2001). By substitutions on the functional group in the 3β-position, chemical diversity of triterpenoids is increased enormously with methyl ethers and acetates as well-known derivatives (Smith and Martin-Smith 1978; Manheim and Mulroy 1978). In monocots, triterpene methyl ethers were identified as common compounds especially in Graminae and Palmae (Garcia et al. 1995; Goh et al. 1988; Russel et al. 1976; Smith and Martin-Smith 1978; Bryce et al. 1967) with cylindrin and lupeol methyl ether as the major compounds for Butia capitata and Orbignya spp. (Garcia et al. 1995). From leaves of O. phalerata, also the saturated derivative lupane methyl ether (3β-methoxy lupane) could be extracted in minor amounts. Recently, lupeol methyl ether was identified for the first time in leaf cuticular waxes also of two C. nucifera ecotypes (Escalante et al. 2002). For the EAT1011×RIT0710 mapping population analyzed in the present study, we found lupeol methyl ether to be the major component and detected minor amounts of lupane methyl ether. Additionally, we found lupeol and its acetate ester to be present in waxes of C. nucifera. These compounds were previously described as constituents of cuticular waxes from R. communis, Euphorbia sp., and Tilia sp. (Guhling et al. 2006; Gülz et al. 1988; Hemmers and Gülz 1988).
Besides this well-known triterpenoid structures in the cuticular waxes of the EAT1011×RIT0710 mapping population, a set of methyl ethers could be identified that derive from cycloartanol type triterpenoids never found in cuticular waxes before. Originally, one of these structures was identified in whole plant extracts of the rutaceen Skimmia wallichii as skimmiwallin (Kostova et al. 1996). Additionally, by spectroscopic analyses, an isomeric form, isoskimmiwallin, was characterized in the leaf waxes of C. nucifera (Escalante et al. 2002). Further detailed analyses detected also the corresponding parent metabolites skimmiwallinol and isoskimmiwallinol as well as there acetylated derivatives already described for the lupeol group.
Trait variation and QTL analyses
The current study on QTLs for epicuticular wax content and composition for the EAT1011×RIT0710 mapping population takes into account several aspects: firstly, wax analysis has been performed on a population of individuals growing under the same environmental conditions rather than on individual trees from different locations and environments which might influence wax content and composition. Secondly, all samples were taken on the same day, thus, minimizing environmental effects. Furthermore, this population represents the two identified groups of coconut germplasm (Rohde et al. 1995; Teulat et al. 2000), namely the “Indo-Atlantic” group (EAT parent) and the “Pacific” group (RIT parent) and, thus, considers a large part of coconut biodiversity.
Some 46 total QTLs and a variable number of individual QTLs were detected for cuticular wax contents and the wax component traits under study. In several cases, QTLs for different traits are colocated or closely linked. These represent probably pleiotropic loci and not closely linked genes for individual traits. The detected correlations and relationships between the different components described above support this hypothesis.
In recent years, the biosynthetic pathways for plant triterpenoids were under intense investigation (for reviews see Abe et al. 1993; Xu et al. 2004; Phillips et al. 2006; Jetter et al. 2006) resulting in a generalized scheme that describes 2,3-oxidosqualene as the last common precursor for most plant triterpenoids. According to this, 2,3-oxidosqualene is cyclized through cationic intermediates by a large family of oxidosqualene cyclase enzymes to triterpene alcohols with the hydroxyl function in the 3β-position (Phillips et al. 2006). On one hand, the most common plant triterpenoids are derived from cyclizations of 2,3-oxidosqualene through the dammerenyl cation leading to, e.g., β-amyrin and lupeol (Kajikawa et al. 2005; Guhling et al. 2006). On the other hand, cyclization through the protosteryl cation leads to lanosterol and cycloartenol with the latter one also being the precursor for membrane sterols, phytohormones, brassinosteroids, and other compounds (Benveniste 2004; Phillips et al. 2006). Whereas the reaction mechanisms and involved enzymes for an increasing number of biosynthetic steps towards the various classes of plant triterpenols could be characterized, information on the methylation or acetylation processes leading to the corresponding methylethers or acetates is still lacking. In the cuticular waxes of the EAT1011×RIT0710 mapping population derivatives of both intermediates were found to be present in substantial amounts.
The analyses of the relative contents of single compounds, on one hand, showed strong negative correlations between the main component LME and all members of the skimmiwallin group (cf. Table 2) indicating a competitive metabolite production by either a lupeol synthase (LUS) or a cycloartenol synthase (CAS). In the last years, both synthase classes from several plant sources have been identified and functionally characterized, e.g., CAS from Arabidopsis thaliana (Corey et al. 1993) and Pisum sativum (Morita et al. 1997), LUS from Betula platyphylla (Zhang et al. 2003) and Olea europea (Shibuya et al. 1999), and both enzyme classes from R. communis (Guhling et al. 2006).
On the other hand, there were strong positive correlations between the methylether (IS and SW) and acetate derivatives (ISA and SWA) of the skimmiwallin group as well as between the both acetate derivatives. This may indicate noncompetitive activities of different enzymes for the catalysis of methylethers and acetates while to our knowledge there is currently no information available on the specificity for the various precursors and the location of the corresponding enzymes which might explain the observed colocations of QTLs.
It will be also necessary to validate the detected QTLs in additional genetic backgrounds. In other progenies, also additional QTLs might be detected, for which the corresponding QTL alleles in our studied population did not cause measurable differences. As pointed out by Baudouin et al. (2006), also the analyses of progenies with more genotypes would be convenient. The increase in available degrees of freedom would permit the application of more sophisticated models involving simultaneous analysis of several QTLs through for example multiple regression models on different intervals. In this way, more precise estimates of QTL effects and locations could be obtained and overestimation of individual QTL effects or even the appearance of minor “false positive” QTLs could be avoided. This approach would be also useful to determine if the detected wax QTLs are stable in different genetic backgrounds and represent independent genes or are due to competing compounds which soften total wax amount differences between genotypes and generate QTL artefacts.
Moreover, in order to determine the contribution of wax amount or specific compounds to disease resistance, it would be necessary to evaluate the incidence of diseases in clonally propagated progeny genotypes. This is a time-consuming and complicated process in coconut.
Alternatively, testcrosses, backcrosses, or even selfings could be derived from genotypes of the studied or other available mapping population and the means of diseased palms could be compared for specific marker allele classes. Also linkage disequilibrium analyses (Jannoo et al. 1999) in less related populations could provide reliable results.
In several cases, SSR markers or even COS clones are located at or nearby QTLs. In the first case, this would imply that selection can take place in different genetic backgrounds, provided that the parental QT allele configuration is known in each case. In the latter case, the gene itself could be located within the corresponding COS clone or at least would facilitate gene identification and cloning through contig building in the surrounding genomic regions.
We have provided a large amount of markers which are useful for coconut and potentially also for oil palm and other related palm species. On one hand, they will serve for the isolation of genes. On the other hand, they provide markers for important characters which can be applied in early selection tests. This will accelerate breeding of coconut and lead to increased productivity of the plantations and improved quality of their products.
References
Abe I, Rohmer M, Prestwich GD (1993) Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem Rev 93:2189–2206
Baker EA (1982) Chemistry and morphology of plant epicuticular waxes. In: Cutler DF, Alvin KL, Price CE (eds) The plant cuticle. Academic, London, pp 139–166
Baudouin L, Lebrun P, Konan JL, Ritter E, Berger A, Billotte N (2006) QTL analysis of fruit components in the progeny of a Rennell Island Tall coconut (Cocos nucifera L.) individual. Theor Appl Genet 112(2):258–268
Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55:429–457
Billotte N, Marseillac N, Risterucci A-M, Adon B, Brottier P, Baurens F-C, Singh R, Herran A, Asmady Billot C, Amblard P, Durand-Gasselin T, Courtois B, Asmono D, Cheah SC, Rohde W, Ritter E, Charrier A (2005) Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacg.). Theor Appl Genet 110:754–765
Bryce TA, Eglinton G, Hamilton RJ (1967) Triterpenoids from New Zealand plants—II. The triterpene methyl ethers of Cortaderia toetoe Zotov. Phytochemistry 6:727–733
Corey EJ, Matsuda SPT, Bartel B (1993) Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc Natl Acad Sci 90:11628–11632
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Escalante F, Gamboa MR, Lecher JG, Arroyo GA, Zizumbo D, Oropeza C, Peña-Rodríguez LM (2002) Major components from epicuticular wax of Cocos nucifera. Rev Soc Quím Méx 46:247–250
García S, Heinzen H, Hubbuch C, Martínez R, De Vries X, Moyna P (1995) Triterpene methyl ethers from Palmae epicuticular waxes. Phytochemistry 39:1381–1382
Goh SH, Lai FL, Gee PT (1988) Wax esters and triterpene methyl ethers from the exocarp of Elaeis guineensis. Phytochemistry 27:877–880
Guhling O, Hobl B, Yeats T, Jetter R (2006) Cloning and characterization of a lupeol synthase involved in the synthesis of epicuticular wax crystals on stem and hypocotyl surfaces of Ricinus communis. Arch Biochem Biophys 448:60–72
Gülz PG, Müller E, Moog B (1988) Epicuticular leaf waxes of Tilia tomentosa and Tilia x europaea L., Tiliaceae. Z Naturfosch 43c:173–176
Gülz PG, Müeller E, Prasad RBN (1991) Developmental and seasonal variations in the epicuticular waxes of Tilia tomentosa leaves. Phytochemistry 30:769–773
Hemmers H, Gülz PG (1988) Triterpenoids in epicuticular waxes of three European Euphorbia species. Z Naturforsch 43c:799–805
Hemmers H, Gülz PG (1989) Tetra- and pentacyclic triterpenoids from epicuticular wax of Euphorbia cyparissias L., Euphorbiaceae. Z Naturforsch 44c:563–567
Jannoo N, Grivet L, Dookun A, D’Hont A, Glaszmann JC (1999) Linkage disequilibrium among sugarcane cultivars. Theor Appl Genet 99:1053–1060
Jeffrey CE (1996) Structure and ontogeny of plant cuticles. In: Kerstiens G (ed) Plant cuticle biosis scientific. BIOS Scientific Publishers, Oxford, pp 33–82
Jenks MA (2002) Critical issues with the plant cuticle’s function in drought tolerance. In: Wood AJ (ed) Biochemical & molecular responses of plants to the environment. Research Signpost, Kerala, India, pp 97–127
Jetter R, Schäffer S (2001) Chemical composition of the Prunus laurocerasus leaf surface. Dynamic changes of the epicuticular wax film during leaf development. Plant Physiol 126:1725–1737
Jetter R, Kunst L, Samuels L (2006) Composition of plant cuticular waxes. In: Riederer M, Müller C (eds) Biology of the plant cuticle. Blackwell, Oxford, pp 144–180
Kajikawa M, Yamoto KT, Fukuzawa H, Sakai Y, Uchida H, Ohyama K (2005) Cloning and characterization of a cDNA encoding b-amyrin synthase from petroleum plant Euphorbia tirucalli L. Phytochemistry 66:1759–1766
Kerstiens G (1996) Cuticular water permeability and its physiological significance. J Exp Bot 47:1813–1832
Knapp SJ, Bridges WC (1990) Programs for mapping quantitative trait loci using flanking molecular markers and nonlinear models. J Heredity 81:234–235
Knapp SJ, Bridges WC, Birkes D (1990) Mapping quantitative trait loci using molecular marker maps. Theor Appl Genet 79:583–592
Kolattukudy PE (1987) Lipid-derived defensive polymers and waxes and their role in plant-microbe interaction. In: Stumpf PK, Conn EE (eds) The biochemistry of plants. Academic, New York, pp 291–314
Kolattukudy PE (1998) Biopolyester membranes of plants: cutin and suberin. Science 208:990–1000
Kostova I, Simeonov M, Iossifova T, Tappe R, Pardeshi N, Budzikiewicz H (1996) Three 9,19-cyclotetracyclic triterpenes from Skimmia wallichii. Phytochemistry 43:643–648
Kurup VVGK, Voleti SR, Rajagopal V (1993) Influence of weather variables on the coconut and composition of leaf surface wax in coconut. J Plant Crops (India) 21:71–80
Lebrun P, Baudouin L, Bourdeix R, Konan JL, Barker JH, Aldam C, Herran A, Ritter E (2001) Construction of a linkage map of the Rennell Island Tall coconut type (Cocos nucifera L.) and QTL analysis for yield characters. Genome 44:962–970
Lemieux B, Koornneef M, Feldmann KA (1994) In: Meyerowitz EM, Somerville CR (eds) Arabidopsis, vol. 27. Cold Spring Harbor Lab, Plainview, New York, pp 1031–1047
Manheim BS, Mulroy TW (1978) Triterpenoids in epicuticular waxes of Dudleya species. Phytochemistry 17:1799–1800
Marcell LM, Beattie G (2002) A Effect of leaf surface waxes on leaf colonization by Pantoea agglomerans and Clavibacter michiganensis. Mol Plant–Microb Interact 15:1236–1244
Markstädter C, Federle W, Jetter R, Riederer M, Hölldobler B (2000) Chemical composition of the slippery epicuticular wax blooms on Macaranga (Euphorbiaceae) ant-plants. Chemoecology 10:33–40
Morita M, Shibuya M, Lee MS, Sankawa U (1997) Molecular cloning of pea cDNA encoding cycloartenol synthase and its functional expression on yeast. Biol Pharm Bull 20:770–775
Phillips DR, Rasbery JM, Bartel B, Matsuda SPT (2006) Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol 9:305–314
Post-Beittenmiller D (1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47:405–430
Ramírez IM, Rodríguez NN, Valdés-Infante J, Capote M, Becker D, Rohde W (2004) Isolation of genomic DNAs from the tropical fruit trees avocado, coconut, guava and mango for PCR-based DNA marker application. Cult Trop 25:33–38
Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52:2023–2032
Ritter E, Salamini F (1996) The calculation of recombination frequencies in crosses of allogamous plant species with application to linkage mapping. Genet Res 67:55–65
Ritter E, Gebhardt C, Salamini F (1990) Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 224:645–654
Rohde W (1996) Inverse sequence-tagged repeat (ISTR) analysis: a novel and universal PCR-based technique for genome analysis in the plant and animal kingdom. J Genet Breed 50:249–261
Rohde W, Kullaya A, Rodriguez J, Ritter E (1995) Genetic analysis of Cocos nucifera L. by PCR amplification of spacer sequences separating a subset of copia-like EcoRI repetitive elements. J Genet Breed 49:179–186
Russell GB, Conner HE, Purdie AW (1976) Triterpene methyl ethers of Chionochloa (Gramineae). Phytochemistry 15:1933–1935
SAS Institute Inc (1989) SAS/STAT users guide version 6, 4th edn. SAS Institute Inc., Cary, N.C, vol. 2
Shibuya M, Zhang H, Endo A, Shishikura K, Kushiro T, Ebizuka Y (1999) Two branches of the lupeol synthase gene in the molecular evolution of plant oxidosqualene cyclases. Eur J Biochem 266:302–307
Smith RM, Martin-Smith M (1978) Triterpene methyl ethers in leaf waxes of Saccharum and related genera. Phytochemistry 17:1307–1312
Sniady V, Becker D, Herrán A, Ritter E, Rohde W (2003) A rapid way of physical mapping in coconut and oil palm. http://www.tropentag.de/2003/abstracts/full/282.pdf pp 1–6
Sommerville C, Browse J, Jaworski JG, Ohlrogge JB (2000) Lipids. In: Buchanan B, Gruissem W, Jones R (eds) Biochemistry and molecular biology of plants. American Society of Plant Phyiologists, Rockville, USA, pp 456–527
Teulat B, Aldam C, Trehin R (2000) An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. Theor Appl Genet 100:764–771
Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Walton TJ (1990) In: Harwood JL, Bowyer JR (eds) Waxes, cutin and suberin. Academic, San Diego, pp 105–158
Xu R, Fazio GC, Matsuda SPT (2004) On the origins of triterpenoid skeletal diversity. Phytochemistry 65:261–291
Yoon R, Alenka HR, Jayakumar P, Dehua L, Dusty PB (1998) Epicuticular wax accumulation and fatty acid elongation activities are induced during leaf development of leeks. Plant Physiol 24:901–911
Zhang H, Shibuya M, Yokota S, Ebizuka Y (2003) Oxidosqualene cyclases from cell suspension cultures of Betula platyphylla var. japonica: molecular evolution of oxidosqualene cyclases in higher plants. Biol Pharm Bull 26:642–650
Acknowledgements
The research was supported, in part, by the European Community under contract ICA4-CT-2001-10066 of the INCO-DEV program and by the Sonderforschungsbereich 567 “Mechanismen der interspezifischen Interaktion von Organismen” of the Deutsche Forschungsgemeinschaft. We thank Fabiola Escalante Erosa and Olga Frank for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Morgante
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Riedel, M., Riederer, M., Becker, D. et al. Cuticular wax composition in Cocos nucifera L.: physicochemical analysis of wax components and mapping of their QTLs onto the coconut molecular linkage map. Tree Genetics & Genomes 5, 53–69 (2009). https://doi.org/10.1007/s11295-008-0168-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0168-7