Abstract
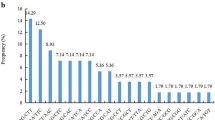
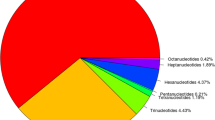
Abundant, codominant simple sequence repeats (SSRs) markers can be used for constructing genetic linkage maps and in marker-assisted breeding programs. Enrichment methods for SSR motifs were optimized with the ultimate aim of developing numerous loci in flowering dogwood (C. florida L.) genome. Small insert libraries using four motifs (GT, CT, TGG, and AAC) were constructed with C. florida ‘Cherokee Brave’ deoxyribonucleic acid (DNA). Colony polymerase chain reaction (PCR) of 2,208 selected clones with three primers we reported previously indicated that 47% or 1,034 of the clones harbored one of the four targeted SSR motifs. Sequencing the putative positive clones confirmed that nearly 99% (1,021 of 1,034) of them contained the desired motifs. Of the 871 unique SSR loci, 617 were dinucleotide repeats (70.8%), and 254 were trinucleotide or longer repeats (29.2%). In total, 379 SSR loci had perfect structure, 237 had interrupted, and 255 had compound structure. Primer pairs were designed from 351 unique sequences. The ability of the 351 SSR primer pairs to amplify specific loci was evaluated with genomic DNA of ‘Appalachian Spring’ and ‘Cherokee Brave’. Of these primers, 311 successfully amplified product(s) with ‘Cherokee Brave’ DNA, 21 produced weak or faint products, and 19 did not amplify any products. Additionally, 218 of the 311 primers pairs revealed polymorphisms between the two cultivars, and 20 out of 218 primers detected an average of 13.7 alleles from 38 selected Cornus species and hybrids. These SSR loci constitute a valuable resource of ideal markers for both genetic linkage mapping and gene tagging of flowering dogwood.

Similar content being viewed by others
References
Ament MH, Windham MT, Trigiano RN (2000) Determination of parentage of flowering dogwood (Cornus florida) seedlings using DNA amplification fingerprinting. J Arboric 26:206–212
Cabe PR, Liles JS (2002) Dinucleotide microsatellite loci isolated from flowering dogwood (Cornus florida L.). Mol Ecol Notes 2:150–152
Caetano-Anollés G, Schlarbaum SE, Trigiano RN (1999) DNA amplification fingerprinting and marker screening for pseudo-testcross mapping of flowering dogwood (Cornus florida L.). Euphytica 106:209–222
Cai HW, Yuyama N, Tamaki H, Yoshizawa A (2003) Isolation and characterization of simple sequence repeat markers in the hexaploid forage grass timothy (Phleum pretense L.). Theor Appl Genet 107:1337–1349
Chen K, Knorr C, Bornemann-Kolatzki K, Ren J, Huang L, Rohrer GA, Brenig B (2005) Targeted oligonucleotide-mediated microsatellite identification (TOMMI) from large-insert library clones. BMC Genetics 6:54 (available at: http://www.biomedcentral.com/1471-2156/6/54)
Cheng HH, Levin I, Vallejo RL, Khatib H, Dodgson JB, Crittenden LB, Hillel J (1995) Development of a genetic map of the chicken with markers of high utility. Poult Sci 74:1855–1874
Connell JP, Pammi S, Iqbal MJ, Huizinga T, Reddy AS (1998) A high throughput procedure for capturing microsatellites from complex plant genomes. Plant Mol Biol Rep 16:341–349
Cornall RJ, Atiman TJ, Hearne CM, Todd JA (1991) The generation of a library of PCR-analyzed microsatellite variants for genetic mapping of the mouse genome. Genomics 10:874–881
Edwards KJ, Barker JHA, Daly A, Jones C, Karp A (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechniques 20:758–760
Ender A, Schwenk K, Stadler T, Streit B, Schierwater B (1996) RAPD identification of microsatellites in Daphnia. Mol Ecol 5:437–441
Fisher D, Bachmann K (1998) Microsatellite enrichment in organisms with large genomes (Allium cepa L.). BioTechniques 24:796–802
Fujishima-Kanaya N, Toki D, Suzuki K, Sawazaki T, Hiraiwa H, Lida M, Hayashi T, Uenishi H, Wada Y, Ito Y, Awata T (2003) Development of 50 gene-associated microsatellite markers using BAC clones and the construction of a linkage map of swine chromosome 4. Anim Genet 34(2):135–141
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and herbaceous plants. Plant Mol Bio Rep 13:207–209
Gardner MG, Cooper SJB, Bull CM, Grant WN (1999) Isolation of microsatellite loci from a social lizard, Egernia stokesii, using a modified enrichment procedure. J Heredity 90:301–304
Hamilton MB, Pincus EL, Di Fiore A, Fleischer RC (1999) Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques 27(3):500–507
Jakse J, Javornik B (2001) High throughput isolation of microsatellites in hop (Humulus lupulus L.). Plant Mol Biol Rep 19:217–226
Jones ES, Dupal MP, Kölliker R, Drayton MC, Forster JW (2001) Development and characterisation of simple sequence repeat (SSR) markers for perennial ryegrass (Lolium perenne L.). Theor Appl Genet 102:405–415
Kandpal RP, Kandpal G, Weissman SM (1994) Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. Proc Natl Acad Sci USA 91:88–92
Kaveriappa KM, Phillips LM, Trigiano RN (1997) Micropropagation of flowering dogwood (Cornus florida L.) from seedlings. Plant Cell Rep 16:485–489
Kijas JMH, Fowler JCS, Garbett CA, Thomas MR (1994) Enrichment of microsatellites from the citrus genome using biotinylated oligonucleotide sequences bound to streptavidin-coated magnetic particles. BioTechniques 16:656–662
Kölliker R, Jones ES, Drayton MC, Dupal MP, Forster JW (2001) Development and characterison of simple sequence repeat (SSR) markers for white clover (Trifolium repens L.). Theor Appl Genet 102:416–424
Lagercrantz U, Ellegren H, Anderson L (1993) The aboundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acid Res 21:1111–1115
Lench NJ, Norris A, Bailey A, Booth A, Markham AF (1996) Vectorette PCR isolation of microsatellite repeat sequences using anchored dinucleotide primers. Nucleic Acid Res 24:2190–2191
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Van De Weg E, Gessler C (2002) Development and characterization of 140 new microsatellites in apple (malus × domestica Borkh.). Mol Breed 10:217–241
Liou JT, Shieh BH, Chen SW, Li C (1999) An improved alkaline lysis method for minipreparation of plasmid DNA. Prep Biochem Biotechnol 29:49–54
Lunt DH, Hutchinson WF, Carvalho GR (1999) An efficient method for PCR-based isolation of microsatellite arrarys (PIMA). Molec Ecol 8(5):891–894
Marinoni D, Akkak A, Bounous G, Edwards K, Botta R (2003) Development and characterization of microsatellite markers in Castanea sativa (Mill.). Mol Breed 11:127–136
Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon AF, Decroocq S (2005) Development and characterization of a larger set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol Breed 15:349–366
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Orton ER Jr (1993) New large-bracted dogwoods from Rutgers University. Comb Proc Intl Plant Propag Soc 43:487–490
Ostrander EA, Jong PM, Rine J, Duyk G (1992) Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. Proc Natl Acad Sci USA 89:3419–3423
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of AFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Rossetto M, McLauchlan A, Harriss FCL, Henry RJ, Baverstock PR, Lee LS, Maguire TL, Edwards KJ (1999) Abundance and polymorphism of microsatellite markers in the tea tree (Melaleuca alternifolia, Myrtaceae). Theor Appl Genet 98:1091–1098
Saal B, Wricke G (1999) Development of simple sequence repeats in rye (Secale cereale L.). Genome 42:964–972
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. CSH Laboratory, Cold Spring Harbor, NY
Smith NR, Trigiano RN, Windham MT, Lamour KH, Finley LS, Wang X, Rinehart TA (2007) AFLP markers identify Cornus florida cultivars and lines. J Am Soc Hortic Sci 132:90–96
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Vadopalas B, Bentzen P (2000) Isolation and characterization of di-and tetranucleotide microsatellite loci in geoduck clams, Panopea abrupta. Molec Ecol 9:1435–1436
Waldbieser GC, Quiniou SM, Karsi A (2003) Rapid development of gene-tagged microsatellite markers from bacterial artificial chromosome clones using anchored TAA repeat primers. BioTechniques 35(5):976–979
Wang XW, Kaga A, Tomooka N, Vaughan DA (2004) The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi]. Theor Appl Genet 109:352–360
Wang XW, Trigiano RN, Windham MT, DeVries RE, Scheffler BE, Rinehart TA, Spiers JM (2007) A simple PCR procedure for discovering microsatellites from small insert libraries. Mol Ecol Notes 7:558–561
Weber JL (1990) Informativeness of human (dC-dA)n-(dG-dT)n polymorphisms. Genomics 7:524–530
Witte WT, Windham MT, Windham AS, Hale FA, Caltterbuck WK (2000). Dogwood for American gardens. The University of Tennessee Agriculture Extension Service, Knoxville. PB1670
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002) Simple sequence repeats for genetic analysis in pear. Euhytica 124:129–137
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Acknowledgments
This work was supported by USDA Agreement no. 58-6404-2-0057. We thank Mary Duke, Linda Ballard, and Xiaofen Liu for library sequencing. Any library/sequence information requirements can be addressed to Dr. Robert N. Trigiano at rtrigian@utk.edu. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Morgante
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
SSR primers developed for flowering dogwood (Cornus florida ) (DOC 697 KB)
Rights and permissions
About this article
Cite this article
Wang, X., Trigiano, R., Windham, M. et al. Development and characterization of simple sequence repeats for flowering dogwood (Cornus florida L.). Tree Genetics & Genomes 4, 461–468 (2008). https://doi.org/10.1007/s11295-007-0123-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-007-0123-z