Abstract
Background
In the last few years, microsatellites have become the most popular molecular marker system and have intensively been applied in genome mapping, biodiversity and phylogeny studies of livestock. Compared to single nucleotide polymorphism (SNP) as another popular marker system, microsatellites reveal obvious advantages. They are multi-allelic, possibly more polymorphic and cheaper to genotype. Calculations showed that a multi-allelic marker system always has more power to detect Linkage Disequilibrium (LD) than does a di-allelic marker system [1]. Traditional isolation methods using partial genomic libraries are time-consuming and cost-intensive. In order to directly generate microsatellites from large-insert libraries a sequencing approach with repeat-containing oligonucleotides is introduced.
Results
Seventeen porcine microsatellite markers were isolated from eleven PAC clones by t argeted o ligonucleotide-m ediated m icrosatellite i dentification (TOMMI), an improved efficient and rapid flanking sequence-based approach for the isolation of STS-markers. With the application of TOMMI, an average of 1.55 (CA/GT) microsatellites per PAC clone was identified. The number of alleles, allele size distribution, polymorphism information content (PIC), average heterozygosity (HT), and effective allele number (NE) for the STS-markers were calculated using a sampling of 336 unrelated animals representing fifteen pig breeds (nine European and six Chinese breeds). Sixteen of the microsatellite markers proved to be polymorphic (2 to 22 alleles) in this heterogeneous sampling. Most of the publicly available (porcine) microsatellite amplicons range from approximately 80 bp to 200 bp. Here, we attempted to utilize as much sequence information as possible to develop STS-markers with larger amplicons. Indeed, fourteen of the seventeen STS-marker amplicons have minimal allele sizes of at least 200 bp. Thus, most of the generated STS-markers can easily be integrated into multilocus assays covering a broader separation spectrum. Linkage mapping results of the markers indicate their potential immediate use in QTL studies to further dissect trait associated chromosomal regions.
Conclusion
The sequencing strategy described in this study provides a targeted, inexpensive and fast method to develop microsatellites from large-insert libraries. It is well suited to generate polymorphic markers for selected chromosomal regions, contigs of overlapping clones and yields sufficient high quality sequence data to develop amplicons greater than 250 bases.
Similar content being viewed by others
Background
Almost all of the applied protocols to isolate microsatellites de novo include construction of partial genomic libraries (selected for small insert size) followed by cumbersome screening steps with hybridization probes [2]. Here, we introduce an improved approach called TOMMI (T argeted O ligonucleotide-M ediated M icrosatellite I dentification) to develop microsatellites by straightforward sequencing of clones isolated from large-insert libraries like PAC (P 1-derived A rtificial C hromosome) and BAC (B acterial A rtificial C hromosome) with repeat-containing oligonucleotides. The need to specifically identify and isolate STS-markers from these types of libraries is unquestionable. First, large-insert libraries are predominantly used in animal genetics, e.g. [3, 4], as tools to identify candidate genes or to generate overlapping contigs of chromosomal regions that are associated with quantitative or economic trait loci (QTL or ETL). Secondly, the overall number of microsatellites present in a genome depends mainly on their complexity and size. Assuming a total size of 3 × 109 bp and an estimated frequency of a dinucleotide repeat every 30–50 kb in mammals (as reviewed by [5]), a genome-wide figure of 100,000 microsatellite markers of that kind can be assumed [6]. However, only approximately 1,200 porcine microsatellites have been reported so far [7]. Furthermore, both the total number and the distribution of the loci are still not sufficient to have well-distributed microsatellite coverage throughout the genome or for several chromosomes, e.g. SSC18 [8]. The objective of the present study was the selective generation of microsatellites from PAC-clones, which were prior to STS development isolated from the porcine PAC library TAIGP714 [3] by a three-dimensional PCR screening strategy [9]. Eight of the eleven clones harbored functional or positional candidate genes involved in health, reproduction, production, and regulation, whereas the other three clones have been used in the attempt to construct a PAC contig covering SSC16q11-13 (Table 1).
Results and discussion
Fifteen of the seventeen microsatellites (Table 2) were developed with sequencing primers containing one selective nucleotide at the 3'-end: (CA)8T (S0701, S0703, and S0767), (CA)8A (S0702, S0704, and S0710), (CA)8G (S0705, S0706, S0712, and S0766), (AC)8C (S0709), (AC)8G (S0707 and S0715), (AC)8T (S0708 and S0711). Characterization of microsatellites S0713 and S0714 was only accomplished by an improved discrimination of the PAC clone sequences with sequencing primers further extended at the 3'-end with a second nucleotide [(CA)8AT for S0713 and (CA)8GC for S0714]. The second nucleotide became necessary because the respective clones TAIGP714L02061Q (for S0713) and TAIGP714I23038Q (for S0714) contained additional (CA)8A or (CA)8G primer binding regions or motifs. Contrary, a further extension with three nucleotides at the 3'-ends of the primers did not result in additional microsatellites in any of the PAC clones or was not required. Therefore, we conclude that repeat primers with two 3'-nucleotides next to the repeat motif are sufficient to detect and sequence all repeats potentially present on a large-insert library clone. The results of our isolation strategy also indicate that two sequencing reactions (the reverse sequencing primer was designed based on the obtained sequences) seem to be sufficient in most cases to gain sequence information of high quality to amplify microsatellites (Table 2). Usage of sequencing primers degenerated at the 3'-end proved, however, to be inadequate as no sequence information at all was achieved. Also, to avoid overlapping primary sequences, oligonucleotides that basically extend the dinucleotide repeat at the 3'-end – such as (CA)8C and (AC)8A – are not recommended. TOMMI proved to be an efficient and reliable isolation strategy. Besides new STS-markers, six previously described microsatellites were also detected. Three of these loci, microsatellites S0111 [10], SW742 [11], and SW813 [12], were initially used as probes for the isolation of clones TAIGP714L02061Q, TAIGP714I23038Q, and TAIGP714F10061Q. The other three already described microsatellite sequences reside on TAIGP714C09004Q [GenBank: AJ440949 (repeat location: 3172–3231) and GenBank: AJ440950 (repeat location: 15831–15860 and 16007–16038)]. They were not further considered in this study as they were not regarded as novel. Independently of our effort, two other groups [13, 14] introduced similar sequencing approaches to generate microsatellites from large-insert libraries. There are, however, several differences between our approach and the ones of the other groups in terms of sequence generation and selective amplification of microsatellites. Here, contrary to Waldbieser and colleagues [14] – who used trinucleotide repeat containing primers for sequencing – both gene-specific primers are not 5'-tailed with extra nucleotide stretches to enable either product labeling or to promote alleged non-template adenylation. Fujishima-Kanaya's group [13] used larger repeat compounds contributing to the primer [(CA/GT)(10) instead of (CA/GT)(8)]. Secondly, the sequencing primers consisted generally of three selective nucleotides at the 3'-end adjacent to the repeat motif (e.g. CNA/GVG). There, the first of the three terminal nucleotides was always identical with the starting nucleotide of the dinucleotide repeat primer used. In addition, primers contained a degenerated base according to the International Union of Biochemistry (IUB) codes at the second position from or directly at the 3'-end. Thirdly, determination of the double-stranded primary DNA sequence stretch was achieved by four sequencing reactions using both a CA-repeat containing primer plus a GT-repeat containing primer heading in the opposite direction and two reverse primers were developed based on the obtained sequence. Finally, they always designed an additional primer pair for the specific amplification of the microsatellite. In contrast, we used the single reverse sequencing primer in combination with a newly developed sequence specific primer (S0766 and S0767) or designed a new primer pair to amplify the microsatellite (S0701 to S0715).
The observed number of alleles per locus (monomorphic locus S0709 is not included in this calculation) in the heterogeneous sampling was as low as 2 (S0702) and as high as 22 (S0713), leading to an average number of 9.94 alleles, NE ranged from 1.05 to 11.54 and both HT and PIC from 0.05 to 0.91 (Table 3).
Due to their isolation from partial genomic libraries selected for small insert sizes most of the publicly available porcine microsatellites lie within DNA-fragments of about 80 to 200 bp. Their potential combination in multiplex assays – also considering different annealing temperatures and technical limitations of the automated sequencers (limited number of available fluorescent dyes) – is therefore hampered. Hence, an enhanced number of genotypes per run can only be achieved by the integration of STS-markers covering a larger allelic spectrum. Thus, we intended and focused on the development of large amplicons for microsatellites by utilizing as much sequence information as possible for primer design. Indeed, fourteen STS-markers had allele sizes of at least 200 bp and for five of the isolated microsatellites, sequence information proved to be good enough to amplify allele sizes of at least 300 bp (Table 3).
By the guided isolation of STS-markers S0709 to S0715 from three SSC16q derived PAC clones (relative position 0 cM to 9.3 cM [7]; 2.33 STS-markers per clone), the marker density in this chromosomal region was improved remarkably. An average of 1.55 new microsatellites was isolated from PAC clones harboring functional candidate genes (S0701-S0708; S0766 and S0767). Considering all used PAC clones and developed STS-markers, 1.55 microsatellites per clone were isolated. As the PAC clones had an average length of 80 kb (as shown by pulsed-field-gel electrophoresis) the frequency of dinucleotide repeats every (30 to) 50 kb [5] was more or less confirmed. TOMMI holds therefore the potential to identify existing STS-markers linked/adjacent to e.g. candidate genes on large-insert library clones. Thus, in combination with a genome scan, respective putative candidate genes could either be transformed to or excluded as positional candidate genes prior to their complete structural characterization including SNP detection. Linkage mapping results for S0701, S0705, S0707, S0711, S0712, S0713, S0715, and S0766 are presented in Table 4. A comparison of their mapping positions with QTL positions (Pig Quantitative Trait Loci (QTL) database [15] reveal that S0705 (64.22 cM), S0707 (43.19 cM), and S0766 (102.50 cM) reside on the respective chromosomes exactly at QTL locations (S0705: backfat between the last 3th and 4th rib; S0707: early growth rate and water holding capacity; S0766: backfat thickness at first rib and intra-muscular fat). The other STS-markers are located in QTL spans of ± 5 cM. This indicates their immediate potential to further dissect these respective QTL regions.
Conclusion
The sequencing strategy described in this study provides a targeted, inexpensive and fast method to develop microsatellites from large-insert libraries. It is also well suited to generate polymorphic markers for selected chromosomal regions and contigs of overlapping clones and yielded sufficient high quality sequence data to develop marker amplicons greater than 250 bases.
Methods
PAC clone isolation and physical mapping
Prior to STS development, a total of 11 clones were isolated from the porcine PAC library TAIGP714 [3] by a three-dimensional PCR screening strategy. PAC-DNA preparations were done according to the manufacturer's protocol (Qiagen, Hilden, Germany). The physical assignment of the PAC clones was performed by Fluorescence in situ Hybridization (FISH) as described in [16] or alternatively by analysis of the INRA-UMN porcine radiation hybrid (IMpRH) panel [17]. Microsatellite primers (Table 3) were used to RH map S0703, S0704 and S0708 – S0715. Marker assignment of S0701, S0702, S0705 – S0707, S0766 and S0767 was performed with primers from further sequence segments of the PAC clones.
Microsatellite generation and characterization
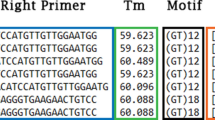
All sequencing reactions and the separation of microsatellites were performed on an ABI PRISM® 3100 DNA analyzer (ABI, Weiterstadt, Germany). Sequencing reactions were done using the BigDye™ Terminator (v 3.0) Cycle Sequencing Kit (ABI, Weiterstadt, Germany). DNA sequencing was performed using 10 pmol of the respective oligonucleotide, 1 μl BigDye Premix and 50–100 ng of purified plasmid DNA as template in a total volume of 10 μl. Sequencing conditions were 96°C for 30 s followed by 30 cycles of 96°C for 10 s, the respective annealing temperature for 5 s and 60°C for 4 min. The optimal annealing temperature for the repeat containing primer was between 50°C and 52°C, except for the generation of sequences for S0714, which were at 56°C. To generate STS-markers, oligonucleotides containing repeat motifs (CA)8 respectively (AC)8 at the 5'-end and few (one or two) non-repetitive bases at the 3'-end were originally used as sequencing primers. Based on the obtained sequence, specific primers were developed and used as reverse oligonucleotides to determine the composition of the repeat region and its 5'-flanking region (Table 2; Figure 1). BLAST comparison followed sequence determination to verify the novelty and uniqueness of the obtained sequences. Depending on the quality of the sequenced stretch, primers were developed to amplify seventeen STS-markers (S0701 to S0715; S0766 and S0767; Table 3). To confirm the sequence identity of the respective microsatellites [GenBank: AY253989 to AY254003, AY731063, and AY731064] on genomic DNA, the resulting PCR products were subcloned into the polylinker of the pGEM®-T vector (Promega, Mannheim, Germany) and three independent clones each were bi-directionally sequenced using standard sequencing primers SP6 (5'-ATT TAG GTG ACA CTA TAG AA-3') and T7 (5'-TAA TAC GAC TCA CTA TAG GG-3').
Evaluation of microsatellites and size determination of alleles were done with appropriate ABI-softwares GENESCAN (3.7) and GENOTYPER (3.6) using GENESCAN™-500ROX™ as internal size standard. Oligonucleotides were designed with the Oligo Selection Program [18] and synthesized by MWG Biotech (Ebersberg, Germany). To characterize size range, number of alleles, polymorphism information content (PIC), average heterozygosity (HT) and effective allele number (NE) of the microsatellites, STS-markers were separately amplified. PCR assays were performed at 54°C for S0706, S0708, S0712, S0713, S0714, and S0767, at 56°C for S0701, S0702, S0703, S0705, S0707 and S0715, and at 58°C for S0704, S0709, S0710, S0711, and S0766 in a RoboCycler Gradient 96® (Stratagene, LaJolla, USA) using PURE Taq Ready-To-Go PCR Beads® (Amersham Biosciences, Freiburg, Germany), along with the respective oligonucleotides (one labeled at the 5'-end alternatively with fluorescent dyes FAM, JOE or NED) and 50 ng of genomic porcine DNA in a volume of 12.5 μl (the concentration of each dNTP is 100 μM in 10 mM Tris-HCl (pH 9.0 at room temperature), 50 mM KCl and 1.5 mM MgCl2). In total, 336 unrelated pigs representing nine European breeds (9 Angeln Saddleback, 18 Bunte Bentheimer, 9 German Edelschwein, 15 German Landrace, 30 Hampshire, 27 Göttingen Minipig, 31 Pietrain, 12 Swabian-Haellian Swine, and 7 European Wild Boar), and six Chinese breeds (30 Chinese Jiangquhai, 28 Chinese Luchuan, 30 Chinese Minpig, 30 Chinese Rongchang, 30 Chinese Tibetan, and 30 Chinese Yushanhei) were investigated. The standard PCR profile was as follows: pre-denaturation at 92°C for 2 min, followed by 35 cycles of 92°C for 30 s, the optimal annealing temperature for 30 s, and 72°C for 30 s. The final cycle had an extension at 72°C for 10 min. PIC, HT and NE were estimated based on algorithms as introduced by Botstein and colleagues [19], Nei [20], and Kimura and Crow [21].
Linkage mapping of STS-markers on the USDA-MARC linkage map
Seven families of the MARC Swine Reference Population were genotyped as described [22]. Amplified DNA was radioactively labeled, separated by denaturing polyacrylamide gel electrophoresis and visualized with autoradiography. To ensure accurate sizing and discrimination of alleles, amplification primers were redesigned to yield smaller products for all markers except S0706, S0707 and S0709. S0767 was not tested in this population. Four markers were not informative in the MARC Swine Reference Population (S0702, S0706, S0709 and S0714) and four primer sets failed to produce reliable products (S0703, S0704, S0708 and S0710). Genotypes were determined and entered into the MARC Genome Database. Each marker was initially assigned to a chromosome based on TWOPOINT results of CRIMAP [23], then multipoint linkage analyses determined the final location of each marker. Genotypic data were evaluated with CHROMPIC and corrections made if necessary. The final position reported is based on the current MARC swine linkage map. Amplification primers for the eight successfully mapped markers are presented in Table 4.
References
Chapman NH, Wijsman EM: Genome screens using linkage disequilibrium tests: optimal marker characteristics and feasibility. Am J Hum Genet. 1998, 63 (6): 1872-1885. 10.1086/302139.
Zane L, Bargelloni L, Patarnello T: Strategies for microsatellite isolation: a review. Mol Ecol. 2002, 11 (1): 1-16. 10.1046/j.0962-1083.2001.01418.x.
Al-Bayati HK, Duscher S, Kollers S, Rettenberger G, Fries R, Brenig B: Construction and characterization of a porcine P1-derived artificial chromosome (PAC) library covering 3.2 genome equivalents and cytogenetical assignment of six type I and type II loci. Mamm Genome. 1999, 10 (6): 569-572. 10.1007/s003359901046.
Buitkamp J, Kollers S, Durstewitz G, Welzel K, Schafer K, Kellermann A, Lehrach H, Fries R: Construction and characterization of a gridded cattle BAC library. Anim Genet. 2000, 31 (6): 347-351. 10.1046/j.1365-2052.2000.00675.x.
Hearne CM, Ghosh S, Todd JA: Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992, 8 (8): 288-294. 10.1016/0168-9525(92)90256-4.
Wintero AK, Fredholm M, Thomsen PD: Variable (dG-dT)n.(dC-dA)n sequences in the porcine genome. Genomics. 1992, 12 (2): 281-288. 10.1016/0888-7543(92)90375-3.
USDA-MARC linkage map.
Campbell EM, Fahrenkrug SC, Vallet JL, Smith TP, Rohrer GA: An updated linkage and comparative map of porcine chromosome 18. Anim Genet. 2001, 32 (6): 375-379. 10.1046/j.1365-2052.2001.00782.x.
Cai L, Taylor JF, Wing RA, Gallagher DS, Woo SS, Davis SK: Construction and characterization of a bovine bacterial artificial chromosome library. Genomics. 1995, 29 (2): 413-425. 10.1006/geno.1995.9986.
Ruyter D, Verstege AJ, van der Poel JJ, Groenen MA: Five porcine polymorphic microsatellite markers. Anim Genet. 1994, 25 (1): 53-
Rohrer GA, Alexander LJ, Keele JW, Smith TP, Beattie CW: A microsatellite linkage map of the porcine genome. Genetics. 1994, 136 (1): 231-245.
Alexander LJ, Rohrer GA, Stone RT, Beattie CW: Porcine SINE-associated microsatellite markers: evidence for new artiodactyl SINEs. Mamm Genome. 1995, 6 (7): 464-468. 10.1007/BF00360655.
Fujishima-Kanaya N, Toki D, Suzuki K, Sawazaki T, Hiraiwa H, Iida M, Hayashi T, Uenishi H, Wada Y, Ito Y, Awata T: Development of 50 gene-associated microsatellite markers using BAC clones and the construction of a linkage map of swine chromosome 4. Anim Genet. 2003, 34 (2): 135-141. 10.1046/j.1365-2052.2003.00967.x.
Waldbieser GC, Quiniou SM, Karsi A: Rapid development of gene-tagged microsatellite markers from bacterial artificial chromosome clones using anchored TAA repeat primers. Biotechniques. 2003, 35 (5): 976-979.
Pig Quantitative trait loci (QTL) database.
Chen KF, Beck J, Huang LS, Knorr C, Brenig B: Assignment of the phosphoglycerate kinase 2 (PGK2) gene to porcine chromosome 7q14-q15 by fluorescence in situ hybridization and by analysis of somatic cell and radiation hybrid panels. Anim Genet. 2004, 35 (1): 71-72. 10.1046/j.1365-2052.2003.01066.x.
Yerle M, Pinton P, Robic A, Alfonso A, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie C, Schook L, Milan D, Gellin J: Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet. 1998, 82 (3-4): 182-188. 10.1159/000015095.
Hillier L, Green P: OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991, 1 (2): 124-128.
Botstein D, White RL, Skolnick M, Davis RW: Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980, 32 (3): 314-331.
Nei M: Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978, 89 (3): 583-590.
Kimura M, Crow JF: The Number of Alleles That Can Be Maintained in a Finite Population. Genetics. 1964, 49: 725-738.
Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW, Beattie CW: A comprehensive map of the porcine genome. Genome Res. 1996, 6 (5): 371-391.
Green P, Falls K, Crooks S: Documentation for CRI-MAP, version 2.4. 1990, Washington University, School of Medicine, St. Louis, MO
Beck J, Knorr C, Habermann F, Fries R, Brenig B: Assignment of the beta-glucuronidase (GUSB) gene to porcine chromosome SSC3p16-->p14 by FISH and confirmation by hybrid panel analyses. Cytogenet Genome Res. 2002, 97 (3-4): 277G-10.1159/000066610.
Mueller A, Knorr C, Habermann F, Slanchev K, Zwilling D, Fries R, Brenig B: Assignment of the beta-N-acetylhexosaminidase gene (HEXB) to porcine chromosome SSC2q21-->q22 by fluorescence in situ hybridization and by analysis of somatic cell and radiation hybrid panels. Cytogenet Genome Res. 2003, 101 (2): 178-10.1159/000074176.
Knorr C, Kollers S, Fries R, Brenig B: Assignment of the CALC-A/alpha-CGRP gene (CALCA) to porcine chromosome SSC2p13-->p11 by fluorescence in situ hybridization and by analysis of somatic cell and radiation hybrid panels. Cytogenet Genome Res. 2002, 97 (1-2): 140F-10.1159/000064050.
Knorr C, Uibeleisen AC, Kollers S, Fries R, Brenig B: Assignment of the homeobox A10 gene (HOXA10) to porcine chromosome SSC18q23-->q24 by FISH and confirmation by hybrid panel analyses. Cytogenet Cell Genet. 2001, 93 (1-2): 145-146. 10.1159/000056972.
Gatphayak K, Knorr C, Habermann F, Fries R, Brenig B: Assignment of the porcine hyaluronidase-3 (HYAL3) gene to SSC13-->q21 by FISH and confirmation by hybrid panel analyses. Cytogenet Genome Res. 2003, 101 (2): 178-10.1159/000074181.
Bull L, Jansen S, Habermann F, Fries R, Knorr C, Brenig B: Assignment of the sperm protein zona receptor tyrosine kinase gene (SPRMTK) to porcine chromosome SSC3q11-->q12 by fluorescence in situ hybridization and by analysis of somatic cell and radiation hybrid panels. Cytogenet Genome Res. 2003, 101 (2): 178-10.1159/000074177.
Chen KF, Beck J, Huang LS, Knorr C, Brenig B: Assignment of the phosphoglycerate kinase 1 (PGK1) gene to porcine chromosome Xq12-q13 by fluorescence in situ hybridization and hybrid panel analyses. Anim Genet. 2004, 35 (2): 143-145. 10.1111/j.1365-2052.2004.01092.x.
Acknowledgements
The authors would like to thank A. Siebels for expert technical assistance. This research project was supported by a grant of the Erxleben Research & Innovation Council to B. Brenig (ERIC-BR1959-2001-06).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
KFC conducted the lab work to isolate and characterize S0701 to S0715 and CK to isolate and characterize S0766 and S0767. CK shared manuscript preparation and editing with KFC, supervised KFC's Ph.D. thesis, evaluated microsatellite data, and organized and provided DNA of the European pig breeds. KBK optimized and conducted fragment analysis and was responsible for evaluation of microsatellite data. JR assisted KFC in the beginning of the project. LSH organized DNA of the Chinese pig breeds. GAR conducted linkage mapping of the markers and edited the manuscript. BB proposed the idea, supervised and commented on the project, was responsible for funding and manuscript editing, and acts as head of the research group in Göttingen.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chen, K., Knorr, C., Bornemann-Kolatzki, K. et al. Targeted oligonucleotide-mediated microsatellite identification (TOMMI) from large-insert library clones. BMC Genet 6, 54 (2005). https://doi.org/10.1186/1471-2156-6-54
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-6-54