Abstract
Despite the high abundance and potential toxicity of ferrocyanide in the environment, data on the bioremediation of these complexes in contaminated soils are missing. In this study we isolated forty bacterial species presented in soil highly contaminated with ferrocyanide complexes, originating from a Manufactured Gas Plant (MGP). All bacterial strains were resistant to ferrocyanide (500 mg L−1). Six isolates showed better growth in the presence of ferrocyanide and were able to use it as a sole nitrogen source. One of them was able to assimilate ferrocyanide‐derived nitrogen and carbon. The strains varied in their tolerance to the ferrocyanide. The Minimum Inhibitory Concentration (MIC) values determined in the rich medium ranged from 1400 mg L−1 to 2000 mg L−1 and in all cases were greater than those set on the minimal medium. Molecular analysis revealed that the investigated isolates had the highest similarity to the Bacillus and Rummeliibacillus lineages. Rummeliibacillus was recognized for the first time for its ferrocyanide-degrading potential. Soil samples collected from MGP sites indicated that the overall indigenous population of microorganisms was low. Total cyanide content ranged from 220 mg kg−1 to 346 mg kg−1. Additionally, elevated Pb concentrations and an imbalanced C:N:P ratio were observed. Our study provides new information about the presence of a well-acclimated bacterial community associated with long-term ferrocyanide-contaminated soil. This bacterial community could play an important role in MGP site bioremediation processes and has the potential for application for other bioremediation purposes; however, it is likely limited due to unfavorable environmental conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Soils are the natural resource that provides the basis for plant growth and thus ensure our daily supply of food; furthermore, soils serve as a water filter, a biodiversity hotspot, and a foundation for buildings and entire cities. Soil functioning is the result of the interplay between (micro)biota living in soil, with their abiotic environment. However, intensive anthropogenic (especially industrial) activities that generate toxic by-products and waste materials have caused soil degradation over the last century and perturbation in the functioning of ecosystem services. Proper, but less well-known examples of manmade industrial activities leading to soil and groundwater contamination are Manufactured Gas Plants (MGP), which were built and operated nationally and worldwide. From the middle of the 19th to the middle of the twentieth century, the manufacturing of gas was a major industry worldwide (Wehrer et al., 2011). Almost every city in the United States and Europe had at least one MGP (Stout & Brey, 2019; Wehrer et al., 2011). It is estimated that there were over 50,000 MGP sites across the United States. Another 60,000 were located in Canada, the UK, continental Europe, and Japan (Murphy et al., 2005). The gas manufacturing and purification processes yielded contaminated residues (including petroleum hydrocarbons, polycyclic aromatic hydrocarbons, cyanides, etc.), which were spread in the surrounding areas or used as a filling material. In soils in contact with the atmosphere, the thermodynamically favored cyanide species are the ferricyanide [Fe(CN)6]3−, and ferrocyanide [Fe(CN)6]4−. The toxicity of these compounds is low, but once exposed to UV radiation, they convert into highly toxic and volatile-free cyanides (Dzombak et al., 2006). Considering the potential toxicity of iron-cyanide compounds and the high CN− concentrations, reaching up to 18,000 mg CN− kg−1 (Sut et al., 2012), remediation of MGP sites in the coming decades may prove to be very challenging. This becomes even more significant due to the worldwide intensive and continuous release of Fe-CN complexes from other industries. Nowadays, pollutants from mining, electroplating, and gas purification processes, as well as anti-caking additives in road salts, are sources of continuous cyanide contamination (Razanamahandry et al., 2017).
Bioremediation utilizes living organisms that use contaminations as a source of nutrients for their metabolic activities (Shishir et al., 2019). This approach has proven environmental applicability and efficacy in mitigating cyanide contamination in soil environments as compared to the more expensive and partially successful chemical and physical remediation techniques (Malmir et al., 2022). To date, several reports have described the ability of microorganisms to efficiently convert cyanide to less toxic compounds in aerobic (Nvokoro & Dibua, 2014, Khamar et al., 2015, Cabello et al., 2018) and anaerobic (Chakrabortis & Veeramani, 2006; Novak et al., 2013) conditions. Additionally, despite the high chemical stability of Fe-CN complexes, several studies have shown that they can be degraded by soil microbes as sources of C and N. Luque-Almagro et al., (2005) reported a Pseudomonas pseudoalcaligenes capable of growth on K4[Fe(CN)6] as the only N-source after pre-cultivation under N-free conditions. Gschwendtner et al. (2016) suggested that long‐term exposition to ferrocyanide applied with deicing salts could lead to a predominance of Actinomycetales, which uses ferrocyanide as a C source. Dimitrova et al. (2020) illustrate the degradation of ferrocyanide by two strains that were isolated from Fe-CN-contaminated soil. However, in this research, no DNA sequencing or the identification of major genes driving cyanide degradation was provided. It is also very important to note that microorganisms capable of metabolizing cyanide in its free form may be not very efficient at degrading metal-cyanide complexes.
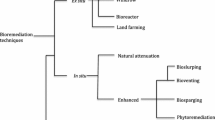
Bioremediation technologies can be categorized as in situ bioremediation and ex-situ bioremediation (Hatzinger et al., 2002). In situ bioremediation is the process that is performed at the original site of the contamination, while the ex-situ bioremediation method involves excavation and transferring contaminated soil to facilitate the degradation of pollutants (Paul et al., 2021). The in situ approach has the significant advantage of avoiding the spread of contaminants, which can occur during transport; it is relatively simple to apply and requires minimal technological equipment (Maitra, 2018). In situ bioremediation is based on the degradative activities of native microbial populations and the premise that the microorganisms present in a contaminated site have developed adaptive mechanisms and are able to degrade contaminations. However, the adapted microorganisms only run the degradation process in suitable environmental conditions (Alori et al., 2022).
Hence, the success of bioremediation (including iron-cyanide bioremediation) depends predominantly upon the presence of microorganisms with the physiological and metabolic potential for survival and the degradation of complex pollutant as well as optimal rates of other various factors – mainly an adequate supply of rate-limiting nutrients (C, N, and P), temperature, pH value and the presence of other contaminants (Azubuike et al., 2016). What is more, in searching for effective candidates for iron-cyanide bioremediation micro-organisms need to be selected not only on their ability to degrade cyanide compounds but also to tolerate the additional contaminations and stresses. Taking into consideration the common occurrence of iron-cyanide in the soil environment and its complex nature, investigating and developing suitable bioremediation strategies using native species is of crucial importance (Kumar et al., 2017). To our knowledge, no study has been conducted to isolate and identify native species present at former MGP sites capable of direct degradation of iron-cyanide complexes as an energy source. Our understanding of the possibilities for in situ bioremediation of former MGP sites is also very limited.
Our research was designed to address these gaps in knowledge by isolating and identifying native bacterial species presented in soil highly contaminated with iron-cyanide complexes, originating from a former MGP site in Cottbus (Germany). Subsequently, the selected strains were cultivated to study minimum inhibitory concentration (MIC) and their potential for ferrocyanide degradation and utilization as a source of nutrients for their metabolic activities. Furthermore, the selected soil properties were investigated to assess the potential for in situ bioremediation.
2 Materials and Methods
2.1 Investigation Site and Soil Sampling
The investigation site is located in the central south-western part of Cottbus (51°45,161’N; 14°18,529’E), in the south of Brandenburg State, in Germany, and it covers an area of 2500 m2 (Fig. 1). Up to 50 cm in depth, the soil is composed of coal, slag and gas purification wastes. In the middle of the twentieth century, the production of manufactured gas was stopped, which resulted in the cessation of the Cottbus MGP. The soil pH varies between 3.2 and 7.7, and the groundwater table is situated at a depth of about 7 m below the surface. The highest cyanide contents were measured in the central part of the site (up to 1600 mg kg−1) (Sut-Lohmann & Raab, 2017). Soil samples at point A-21 were collected at a depth of 20 cm and 30 cm using a hand driller.
2.2 Media and Growth Conditions
Bacteria were cultivated in Bunt and Rovira (BR) medium (glucose 20.0 g L−1; peptone 1.0 g L−1; yeast extract 1.0 g L−1; (NH4)2 SO4 0.5 g L−1; K2HPO4 0.4 g L−1; MgCl2 0,1 g L−1; FeCl3 0.01 g L−1; filtered soil extract 250 ml L−1; pH 7.0; agar 20.0 g L−1) and M9 minimal medium (pH 7) with glucose (5 g L−1) as a carbon source and ammonium chloride (1 g L−1) as a nitrogen source or potassium ferrocyanide K4[Fe(CN)6] x 3H2O (1 mM) as a carbon and/or nitrogen source. Fungi were cultivated on Martin’s medium (glucose 10.0 g L−1; peptone 5.0 g L−1; K2HPO4 1.0 g L−1; MgS04 × 7H20 0.5 g L−1; rose bengal 1:300 solution 10.0 ml L−1) with antibiotics. Media were autoclaved for 20 min at 15 psi and 121 °C. Filter-sterilized (0.2-mm-pore-size filter) solutions of K4[Fe(CN)6] x 3H2O were added to the BR and M9 medium. Filter-sterilized (0.2-mm-pore-size filter) streptomycin (30 mcg ml−1) was added to the Martin’s medium. Incubation was performed in the dark, at 25 °C and with an agitation of 350 rpm. The cultures were inoculated into the broth medium with a 1% (v/v) inoculum.
2.3 Microbial Counts
10 g of soil samples were added to 90 mL of sterile physiological salt solution (0.9% NaCl). After homogenization (350 rpm for 30 min), this solution was decimally diluted (10−1 to 10−7), and aliquots of the resulting solutions were plated on appropriate culture media. After incubation for up to 5 days, the colony-forming units (CFU) were counted. The total number of bacterial CFU was estimated on Bunt and Rovira (BR) medium. The total number of fungal CFU was estimated on Martin’s medium.
2.4 Isolation of Bacterial Strains
For bacterial isolation purposes, 10 g of the soil sample was dissolved in 90 ml of sterile physiological salt solution (0.9% NaCl). After homogenization (350 rpm for 30 min) a serial dilution in a sterile physiological salt solution (0.9% NaCl) up to 10−4 was prepared and an aliquot of 0.1 ml was spread over Bunt and Rovira (BR) medium agar plates. The plates were incubated at 25 °C for 48 h. After several replications of plating, forty morphologically different colonies were isolated (marked as MGP-1 to MGP-40). All isolated strains were stored in glycerol at − 80 °C, creating a bacterial culture collection.
2.5 Bacterial Growth Inhibition/Stimulation Tests
For the purpose of a strains comparison study, growth inhibition/stimulation was determined after 48 h of incubation in a BR broth with a ferrocyanide concentration of 500 mg L−1. After the incubation time, OD600 of the cultures was measured. All experiments were carried out in triplicate, with appropriate controls. Inhibition/stimulation of bacterial growth by the ferrocyanide was calculated according to the following formula: 100% ( OD600 bacteria grown in the presence of cyanide—OD600 control)/OD600 control.
2.6 Bacterial Ability to Use Ferrocyanide as a Nitrogen and/or Carbon Source
Based on the bacterial growth inhibition test, six bacterial strains were selected to assess their ability to use ferrocyanide as a nitrogen and/or carbon source. Particulate bacterial strains were inoculated into a 100-ml Erlenmeyer flask containing 50 ml of the M9 minimal medium with (i) glucose as a carbon source and ammonium chloride as a nitrogen source, (ii) glucose as a carbon source and potassium ferrocyanide K4[Fe(CN)6] x 3H2O as a nitrogen source and (iii) potassium ferrocyanide K4[Fe(CN)6] x 3H2O as a carbon and nitrogen source. After 14 days of incubation, OD600 of the cultures was measured.
2.7 MIC Tests
To identify the concentration of ferrocyanide, which inhibits the growth of selected bacterial strains, MIC tests were performed. A concentration series of 400 mg L−1 to 2200 mg L−1 (200 mg L−1 increment) of potassium ferrocyanide K4[Fe(CN)6] x 3H2O in BR medium and M9 medium was used to determine MIC. An overnight culture was inoculated into the liquid medium and OD600 was measured at 24, 48, and 72 h. MIC was described as the lowest concentration of potassium ferrocyanide K4[Fe(CN)6] x 3H2O, where bacterial growth after 72 h incubation time was not observed.
2.8 Bacterial Strains Identification
Bacterial colonies (six strains) showing vigorous growth in the presence of potassium ferrocyanide were propagated, then genomic DNA was extracted using a DNeasy PowerSoil kit (QIAGEN). The soil samples were lysed and subjected to inhibitor removal for cleanup. Following the cleanup, the purified lysate was mixed with DNA binding solution and passed through a silica membrane. The membrane was washed with a two-step washing regime. DNA was finally eluted using a proper elution buffer. Approximately 100 ng of DNA was used as the template for PCR amplification of nearly full-length bacterial 16S rRNA gene fragments using a complete PCR kit with Taq polymerase (A&A Biotechnology) and the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The reactions were performed in a PTC-200 thermal cycler (MJ Research, Inc., USA) under the following conditions: 95 °C for 5 min, followed by 20 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 90 s; and then 5 cycles of 95 °C for 30 s, 46 °C for 30 s, 72 °C for 1.5 min; with a final extension at 72 °C for 10 min. Amplification products purified with EPPiC Fast (A&A Biotechnology) were directly sequenced using BigDye Terminator v.3.1 chemistry on an ABI3730xl DNA Genetic Analyzer (Applied Biosystems) with the primers F27 and 1492R. The sequencing results were assembled with Geneious assembler software using default parameters, except that the Error Probability Limit was set to 0.005 (Geneious 10.2.3; http://www.geneious.com). The 16S rDNA sequences obtained were then used as queries against the NCBI database using BLAST.
2.9 Determination of Physicochemical Properties of the Soil
The following laboratory analyses of the soil samples were carried out:
-
Cyanide determination using automated spectrophotometrical flow injection system FIA (DIN EN ISO 14 403 D) after 12 h extraction with 1 M sodium hydroxide (1:10, w/v). Iron-cyanide containing solution was prepared using potassium hexacyanoferrate (II) L − 1 (Sigma‐Aldrich)
-
Spectroscopic analysis using a FTIR spectrometer (Bruker, Tensor 27, HTS-XT, resolution 4 cm 1, 40 scans per sample) in the MIR range, applying the diffuse reflectance infrared Fourier spectroscopy (DRIFTS)
-
pH using the potentiometric method in a suspension with water and 1 mol dm−3 solution of KCl
-
Content of total organic carbon (TOC), total nitrogen (N) and total sulfur (S) by dry combustion (Vario MacroCube, Elementar, Germany)
-
Total contents of P, Na, K, Ca, Mg, Fe, Al, Mn, V, Cr, Co, Ni, Cu, Zn, Cd, Pb, As, Li, Sr and Ba using inductively-coupled plasma atomic emission spectrometry (ICP-AES, Avio 200, Perkin Elmer) after the digestion of samples in a mixture of 40% HF, 65% HNO3 and 38% HCl (5:3:2 by volume) using the microwave digestion system EthosUp, Milestone.
2.10 Statistical Analysis
The collected data were subjected to an analysis of variance (ANOVA) for comparison of the means. Significant differences were calculated according to Tukey’s post hoc test at a p < 0.05 significance level.
3 Results
3.1 Microbial Counts
The results presented in Fig. 2 refer to the abundance of bacteria and fungi. Data concerning bacteria (Fig. 2a) showed that their numbers in both soil layers (20 and 30 cm) were comparable. The total number of bacterial CFU was slightly higher at a depth of 20 cm; however, the differences were statistically insignificant. The results obtained for fungi (Fig. 2b) showed higher variability depending on the depth. A significantly lower abundance of fungal CFU was recorded in the soil taken from 30 cm.
3.2 Bacterial Isolation and Growth Inhibition/Stimulation Tests
The selection of bacteria from the native microflora of iron-cyanide-contaminated soil led to the isolation of forty isolates forming a bacterial culture collection (marked as MGP 1 to MGP 40). The results concerning bacterial growth inhibition/stimulation in the presence of ferrocyanide are presented in Fig. 3a and b. The effect of ferrocyanide was tested in assays using the bacterial culture collection. All isolated strains could tolerate a high ferrocyanide concentration (500 mg L−1). The growth of thirty-four isolates was lower in the presence of ferrocyanide compared to the control. Six out of forty isolates (isolate MGP 12, MGP 25, MGP 26, MGP 28, MGP 29 and MGP 31) showed better growth in the presence of ferrocyanide (Fig. 3a). The inhibitory effect of ferrocyanide hesitated between 0,9% to 56%, while its stimulatory effect hesitated between 2 to 49% (Fig. 3b). Results obtained indicate the presence of a well-acclimated iron-cyanide resistant soil bacterial community and suggest that, in some cases, ferrocyanide was an additional N and/or C source.
Comparison of bacterial growth estimated based on OD600 (a) and inhibition/stimulation test estimated in comparison to control samples (b). The measurements were taken in the presence of ferrocyanide (500 mg L-1) after 48 h of incubation in a BR broth in aerobic conditions. The experiment was conducted three times (n = 3). Bars labeled with different letters differ significantly (p < 0.05)
3.3 Bacterial Ability to Use Ferrocyanide as a Nitrogen and/or Carbon Source
Six bacterial strains indicating higher growth in the presence of ferrocyanide were selected to assess their ability to use ferrocyanide as a nitrogen and/or carbon source (Fig. 4). All the tested strains were able to grow in a minimal M9 medium with glucose as a carbon source and ferrocyanide as a nitrogen source. The growth rate was species-dependent. These results show that the strains under examination can obtain their nitrogen requirement from ferrocyanide. Additionally, isolate MGP 31 was able to use potassium ferrocyanide as a nitrogen and carbon source. All strains investigated grew in M9 minimal medium with glucose as a carbon source and ammonium chloride as a nitrogen source and did not grow in the nitrogen-free control medium or the carbon-free control medium. Together, these results demonstrate that isolates MGP 12, MGP 25, MGP 26, MGP 28, MGP 29 and MGP 31 have great biotechnological potential.
Growth curves of six bacterial isolates cultivated in a presence of ferrocyanide as a nitrogen and/or carbon source (n = 3): isolate MGP 12 (a); isolate MGP 25 (b); isolate MGP 26 (c); isolate MGP 28 (d); isolate MGP 29 (e); isolate 31 (f); Control – minimal medium with glucose as a carbon source and ammonium chloride as a nitrogen source; Fe-CN—N – minimal medium with glucose as a carbon source and potassium ferrocyanide as a nitrogen source; Fe-CN – N/C – minimal medium with potassium ferrocyanide as a carbon and nitrogen source
3.4 MIC Tests
The MIC tests of the six strains on BR and M9 media are presented in Table 1. The strains that were examined differed in their tolerances to the ferrocyanide, with MGP 12, MGP 25, MGP 28 and MGP 31 being more tolerant, evidenced by higher MIC values (1800 mg L−1 to 2000 mg L−1) than MGP 26 and MGP 29 (MIC 1400 mg L−1 for both). Additionally MIC values determined in the rich BR medium were always greater than those in the M9 medium. The greatest disparities were observed for the MGP 25 and MGP 31 strains, while the least were for MGP 12 and MGP 29.
3.5 Identification of Bacterial Strains
Molecular analysis based on 16S rRNA gene sequencing showed that isolates MGP 12, MGP 25, MGP 28, MGP 29, and MGP 31 had the highest similarity to the Bacillus strains, while the isolate MGP 26 was classified in the Rummeliibacillus lineage (Fig. S1). Strain MGP 12 belonged to the Bacillus pumilus-safensis-zhangzhouensis-australimaris clade, and the sequence fragment we analyzed (1 441 bp) showed 100% similarity to the type strains of B. pumilus (ATCC 7061 T) and Bacillus safensis (MS11T). Strain MGP 25 was almost 98% similar to the strain DRG5 of Bacillus foraminis. Strain MGP 28 was classified in the Bacillus megaterium-aryabhattai-zanthoxyli-oryzaecorticis-ginsengisoli clade, showing 100% similarity to some strains of these species. The 16S rRNA sequence of isolate MGP 29 was identical to the sequences of the Bacillus simplex, Bacillus huzihouensis, Bacillus choshinensis, and Bacillus frigoritolerans strains, including the strain type DSM 8801 T of B. frigoritolerans previously described as Brevibacterium frigoritolerans. Similarly, strain MGP 31 showed 100% similarity to the Bacillus cereus-thuringiensis-toyonensis-mobilis clade. Since the 16S rDNA gene sequence does not sufficiently distinguish between species belonging to the Bacillus genera, as also confirmed by our phylogenetic analysis, it is advisable to use the gyrase subunit B gene (gyrB gene) for the identification of species belonging to the Bacillus genus. A detailed MLSA analysis of the Bacillus isolates obtained will therefore be performed in the next step of our research. Only one strain, MGP 26, belonged to the Rummeliibacillus genus, to the Rummeliibacillus pycnus clade, showing 100% similarity to the Rummeliibacillus sp. 18JY16-4 strain described in South Korea (NCBI dataset).
3.6 Soil Characteristics
The Mid Infrared spectra of the analyzed soil samples collected from depths of 20 cm and 30 cm are shown in Fig. 5, indicating the presence of the ferric ferrocyanide, with the stretching vibration ranging from 2092 to 2084 cm−1. The total CN concentration were 220 mg kg−1 and 346 mg kg−1 for a depth 20 cm and 30 cm, respectively. The soil samples collected from depths of 20 cm and 30 cm were comparable in terms of pH (close to neutral) and contents of Na, Ca, Mg, Al and Zn. Soil taken from 20 cm contained more TOC, N, S, Cu, Pb, Sr and Ba, whereas soil from 30 cm had more P, K, Fe, Mn, Ti, Zr, Ni, V and Cr (Table 2). TOC:N:P values were 45.2:3.3:1 and 17.8:1:1.6, respectively. Generally, both soils contained typical amounts of mineral, except Pb, the concentrations of which were elevated, indicating anthropogenic contamination. The former MGP site is characterized by the contaminated purifier waste material that originates from the coal gas purification process and is present at the site between 20–100 cm depth (Sut-Lohmann et al., 2015). This very inhomogeneous waste layer was composed of impure coal gas, tar, hydrogen sulfide (H2S), ammonia, (NH3), hydrocarbons (PAH) hydrogen cyanide (HCN), and various heavy metals. It is suggested that in the past, contaminated purifier wastes were used as a filling material and exposed directly to soil. This would explain the difference in the elemental concentration between 20 and 30 cm sampling depth.
4 Discussion
A wide range of pollutants reach the soils of natural and managed ecosystems. These chemicals, which include agriculture pollutants, heavy metals, acid deposition, and a range of industrial chemicals, can affect soil's physical, chemical, and biological functions (Edwards, 2002; Zarime et al., 2023). Microbial abundance in the soil is strongly dependent on soil properties but also extremely site-specific in every soil environment. It has been estimated that 1 g of soil can harbor approximately 108–1010 bacterial cells and approximate 200 m fungal hyphae (Xiong & Lu, 2022). A microbiological examination of soil from a former MGP site in Cottbus showed that the indigenous population of microorganisms was low (32 – 38 × 103 bacterial CFU/g of soil and 7.3 – 18 × 101 fungal CFU/g of soil). Microbial, especially fungal abundance was also related with total CN concentration. These results supported that the adaptation of microbial communities to long-term soil pollution decreases the overall microbial population. Our findings suggest that bacteria exhibit greater tolerance to iron-cyanide than fungi, and may play a crucial role in the bioremediation of soil contaminated by cyanide. Even though microbial enumeration is not a direct measure of their activity in soils, it indicates microbial vitality and/or biodegradative potential (Fingerman & Nagabhushanam, 2016). At the former MGP site in Cottbus, despite the high soil contamination in the form of iron-cyanide complexes, petroleum hydrocarbons, and polycyclic aromatic hydrocarbons, sulfur, and heavy metals in increased amounts were found (Sut et al., 2014). High levels of contaminants often inhibit a large sector of the soil microorganisms, whereas soil with lower contamination shows a greater number and diversity of microorganisms (Dean-Ross, 1989). Forsyth et al. (1995) have shown that when the population of indigenous microorganisms is less than 105 CFU/g of soil, bioremediation does not occur at a significant rate.
Microorganisms useful in cyanide bioremediation must have a cyanide-insensitive metabolism. However, the formation of stable metal-cyanide complexes in the soil environment complicates the bioremediation process. In this situation, bacterial proliferation requires additional specific metal uptake systems (organic compounds called siderophores). Cyanotrophic microorganisms also need an assimilatory pathway to use cyanide and its derivatives as a nitrogen or/and carbon source for growth. Thus, the biological assimilation of iron-cyanide requires at least three separate processes, i.e., a cyanide resistance mechanism, a system for metal binding, and a cyanide assimilation pathway (Luque-Almagro et al., 2005). In this study from the forty ferrocyanide-resistant (500 mg L−1) isolated bacteria, six strains were able to use ferrocyanide as a nitrogen source, and one strain was able to use ferrocyanide as a nitrogen and carbon source. This capability, in combination with the high resistance to cyanide (MIC up to 2000 mg L−1), makes this strain a very good candidate for future metal-cyanide bioremediation treatment.
Molecular analysis based on 16S rRNA gene sequencing showed that six out of the forty isolates had the highest similarity to the Bacillus and Rummeliibacillus lineage. Among the bacterial genera, the members of the genus Bacillus have considerable potential to degrade or transform various toxic substances (Arora, 2020; Harirchi et al., 2022). The genus Bacillus belongs to the Bacillaceae family, which comprises 293 species and subspecies (Patel & Gupta, 2020). So far, the role of various Bacilli in the biodegradation of dyes, pesticides, herbicides, chlorophenols, nitrophenols, chloronitrophenols, drugs, explosives, crude oil waste, plastics, alkaline lignin, and other natural compounds have been well characterized (Arora, 2020). The bioremediation of a cyanide contaminated environment by the Bacillus genus has also been described. Mekuto et al. (2013) have shown that mixed culture dominated by Bacillus sp. (Bacillus safensis, Bacillus lichenformis and Bacillus tequilensis) was able to tolerate and biodegrade the cyanide in cultures containing 200 and 400 mg CN L−1. Kandasamy et al. (2015) described the efficient cyanide bioremediation and high microbial growth obtained with Bacillus anthracis, Bacillus pumilus and Bacillus weihenstephenensis. Guadalima and Monteros (2018) evaluated the effect of rotational speed and carbon source on the biological removal of cyanide present on gold mine wastewater, using a Bacillus sp. Wu et al. (2014) have shown that Bacillus sp. CN-22 can tolerate cyanide concentration of 700 mg L−1 and achieve degradation of CN − at 96.69%. Javaheri Safa et al. (2017) found that, Bacillus sp. M01 PTCC 1908 has high resistance (15 mM–976 mg L−1) and degrading ability (10 mM–562 mg L−1) and suggested using these bacteria for bioremediation processes in many industries that deal with cyanide contamination. Additionally, many Bacilli have been characterized for the remediation of heavy metals (Ayangbenro & Babalola, 2020; Njoku et al., 2020; Sher et al., 2021). Rummeliibacillus application potential has been highlighted in biotechnology (Li et al., 2019); however, it has not yet been described in the context of cyanide or metal-cyanide bioremediation.
Soil pH plays a vital role in the activity of soil microorganisms. It affects a range of soil biological properties, including bioremediation. Generally, a pH value of 5.5 to 8 is needed for optimal microbial activity (Neina, 2019). Previous studies on the biodegradation of ferrocyanide have determined an optimum range for the pH value in the biodegradation of ferrocyanide, ranging from 5.5 to 6.5 (Dash et al., 2008). In contrast, Pseudomonas pseudoalcaligenes CECT5344 is able to grow by using ferrocyanide as the sole nitrogen source under alkaline conditions with an optimal pH value of 9 (Huertas et al., 2010). However, considering the optimum pH for the growth of most cyanide-degrading bacteria and for cyanide-degrading enzymes, the soil tested seems to be optimal in terms of this parameter.
Bioremediation can be effective only where environmental conditions permit microbial growth and activity. Carbon (C), nitrogen (N) and phosphorus (P) are crucial for this process. The recommended C: N: P ratio for enhanced biodegradation processes commonly cited as optimal is in the range of 100:10:1 to 100:1:0.5 (Leys et al., 2005; Ouriache et al., 2020). The results showed that the C: N: P ratio in the tested soil was 45.2:3.3:1 and 17.8:1:1.6 (for the depths 20 cm and 30 cm, respectively). It should be noted that cyanide compounds are a better source of N than of C. The oxidation states of C and N in CN− are +2 and −3, respectively, and the oxidation state of N in CN− is identical to that of N in NH4+ (Luque-Almagro et al., 2005). Thus, special attention should be given to the carbon content during cyanide bioremediation. The biostimulation process (addition of limiting nutrients to support microbial growth) and regulation of imbalance among carbon, nitrogen, and phosphorus content in the tested soil can significantly increase the microbial biodegradation process. Additionally, the observation that MIC values depended on the medium composition and increased in the rich medium suggests that optimal growth conditions enhance microbial ferrocyanide resistance.
The soil samples examined exhibited elevated Pb concentrations. The bioremediation process has often been shown to be limited by the presence of heavy metals. They are well-known to inhibit the activity of microbial enzymes as well as disturb carbon, nitrogen and organic matter transformation and reduce microbial biodiversity and biomass (Jarosławiecka & Piotrowska-Seget, 2022). However, there are also several protection mechanisms of heavy metal resistance by microbial cells (Bruins et al., 2000). Up to this point, several species of Bacillus, especially from the cereus and subtilis groups, have been described as being resistant to metal ions. Bacterial strains resistant and able to adsorb arsenic, lead, copper, cadmium, and zinc were usually isolated from heavy metal-contaminated areas (Jeyakumar et al., 2023; Pandey et al., 2013). Based on our research, we can speculate that the structure of microbial communities in the tested soil was shaped not only by the specific iron-cyanide contamination but also by the other co-contaminants. Isolated bacterial strains also exhibited resistance to these co-contaminants.
5 Conclusions
In conclusion, this study is the first to report that long-term contaminated soil from a former MGP site can be a source of a well-established ferrocyanide-resistant bacterial community. Selected members of this community, classified as Bacillus and Rummeliibacillus strains, are able to use ferrocyanide complexes as a nitrogen and carbon source by degrading them. These strains can play an important role in the MGP site in situ bioremediation processes. Additionally, in this study Rummeliibacillus was recognized for the first time for its ferrocyanide-degrading potential. Although more research is needed, these findings can be utilized to develop bio-treatment in many industries that deal with cyanide contamination. This work also underlines the presence of unfavorable environmental conditions on the MGP sites, which have the potential to inhibit microbial abundance and activities, thereby posing a challenge to the success of bioremediation efforts. This information could be applied to develop strategies to optimize the bioremediation of MGP sites all over the world.
Data Availability
The collected data are available as open data via the Warsaw University of Life Sciences online repository: https://doi.org/10.18150/W7IVU3
References
Alori, E. T., Gabasawa, A. I., Elenwo, C. E., & Agbeyegbe, O. O. (2022). Bioremediation techniques as affected by limiting factors in soil environment. Frontiers in Soil Science, 47. https://doi.org/10.3389/fsoil.2022.937186
Arora, P. K. (2020). Bacilli-mediated degradation of xenobiotic compounds and heavy metals. Frontiers in Bioengineering and Biotechnology, 8, 570307. https://doi.org/10.3389/fbioe.2020.570307
Ayangbenro, A. S., & Babalola, O. O. (2020). Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Scientific Reports, 10(1), 1–12. https://doi.org/10.1038/s41598-020-75170-x
Azubuike, C. C., Chikere, C. B., & Okpokwasili, G. C. (2016). Bioremediation techniques–classification based on site of application: Principles, advantages, limitations and prospects. World Journal of Microbiology & Biotechnology, 32, 180. https://doi.org/10.1007/s11274-016-2137-x
Bruins, M. R., Kapil, S., & Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicology and Environmental Safety, 45(3), 198–207. https://doi.org/10.1006/eesa.1999.1860
Cabello, P., Abril, A., & Luque-Almargo, V. (2018). Assimilation of cyanide and cyano-derivatives by pseudomonas pseudoalcaligenes CECT5344: from omic approaches to biotechnological applications. FEMS Microbiology Leters, 365(6), fny032. https://doi.org/10.1093/femsle/fny032
Chakrabortis, S., & Veeramani, H. (2006). Efects of HRT and recycle ratio on removal of cyanide, phenol, thiocyanate, and ammonia in an anaerobic–anoxic–aerobic continous system. Process Biochemistry, 4(1), 96–105. https://doi.org/10.1016/j.procbio.2005.03.067
Dash, R. R., Balomajumder, C., & Kumar, A. (2008). Treatment of metal cyanide bearing wastewater by simultaneous adsorption and biodegradation (SAB). Journal of Hazardous Materials, 152(1), 387–396. https://doi.org/10.1016/j.jhazmat.2007.07.009
Dean-Ross, D. (1989). Bacterial abundance and activity in hazardous waste-contaminated soil. Bulletin of Environmental Contamination and Toxicology, 43(4). https://doi.org/10.1007/BF01701928
Dimitrova, T. (2010). Determination of cyanides in contaminated soils using micro distillation-and spectrophotometric flow injection system. Faculty of environmental sciences and process engineering. Brandenburg University of Technology, 37–43.
Dimitrova, T., Repmann, F., & Freese, D. (2020). Degradation of ferrocyanide by natural isolated bacteria. International Journal of Phytoremediation, 22(1), 20–28. https://doi.org/10.1080/15226514.2019.1633996
Dzombak, D. A., Ghosh, R. S., & Young, T. C. (2006). Physical–chemical properties and reactivity of cyanide in water and soil. In D. A. Dzombak, R. S. Ghosh, & G. M. Wong-Chong (Eds.), Cyanide in water and soil: chemistry, risk and management (57–88). Taylor and Francis/CRC Press. ISBN9780429138959.
Edwards, C. A. (2002). Assessing the effects of environmental pollutants on soil organisms, communities, processes and ecosystems. European Journal of Soil Biology, 38(3–4), 225–231. https://doi.org/10.1016/S1164-5563(02)01150-0
Fingerman, M., & Nagabhushanam, R. (2016). Bioremediation of aquatic and terrestrial ecosystems. Book. 2016/04/19.
Forsyth, J. V., Tsao, Y. M., & Bleam, R. D. (1995). Bioremediation: When is bioaugmentation needed? In R. E. Hinchee, J. Fredrickson, & B. C. Alleman (Eds.), Bioaugmentation for site remediation (pp. 1–14). Battelle Press.
Gschwendtner, S., Mansfeldt, T., Kublik, S., Touliari, E., Buegger, F., & Schloter, M. (2016). Long-term ferrocyanide application via deicing salts promotes the establishment of Actinomycetales assimilating ferrocyanide-derived carbon in soil. Microbial Biotechnology, 9(4), 502–513. https://doi.org/10.1111/1751-7915.12362
Guadalima, M. P. G., & Monteros, D. A. N. (2018). Evaluation of the rotational speed and carbon source on the biological removal of free cyanide present on gold mine wastewater, using a rotating biological contactor. Journal of Water Process Engineering, 23, 84–90. https://doi.org/10.1016/j.jwpe.2018.03.008
Harirchi, S., Sar, T., Ramezani, M., Aliyu, H., Etemadifar, Z., Nojoumi, S. A., & Taherzadeh, M. J. (2022). Bacillales: from taxonomy to biotechnological and industrial perspectives. Microorganisms, 10(12), 2355. https://doi.org/10.3390/microorganisms10122355
Hatzinger, P. B., Whittier, M. C., Arkins, M. D., Bryan, C. W., & Guarini, W. J. (2002). In-situ and ex-situ bioremediation options for treating perchlorate in groundwater. Remediation Journal, 12(2), 69–86. https://doi.org/10.1002/rem.10026
Huertas, M. J., Sáez, L. P., Roldán, M. D., Luque-Almagro, V. M., Martínez-Luque, M., Blasco, R., & García-García, I. (2010). Alkaline cyanide degradation by Pseudomonas pseudoalcaligenes CECT5344 in a batch reactor. Influence of pH. Journal of Hazardous Materials, 179(1–3), 72–78. https://doi.org/10.1016/j.jhazmat.2010.02.059
Jarosławiecka, A. K., & Piotrowska-Seget, Z. (2022). The effect of heavy metals on microbial communities in industrial soil in the area of Piekary Śląskie and Bukowno (Poland). Microbiology Research, 13(3), 626–642. https://doi.org/10.3390/microbiolres13030045
JavaheriSafa, Z., Aminzadeh, S., Zamani, M., & Motallebi, M. (2017). Significant increase in cyanide degradation by Bacillus sp. M01 PTCC 1908 with response surface methodology optimization. AMB Express, 7, 1–9. https://doi.org/10.1186/s13568-017-0502-2
Jeyakumar, P., Debnath, C., Vijayaraghavan, R., & Mutahuraj, M. (2023). Trends in bioremediation of heavy metal contaminations. Environmental Engineering Research, 28(4). https://doi.org/10.4491/eer.2021.631
Kandasamy, S., Dananjeyan, B., Krishnamurthy, K., & Benckiser, G. (2015). Aerobic cyanide degradation by bacterial isolates from cassava factory wastewater. Brazilian Journal of Microbiology, 46, 659–666. https://doi.org/10.1590/S1517-838246320130516
Khamar, Z., Makhdoumi-Kakhki, A., & Gharaie, M. M. (2015). Remediation of cyanide from the gold mine tailing pond by a novel bacterial co-cul ture. International Biodeterioration and Biodegradation, 99, 123–128. https://doi.org/10.1016/j.ibiod.2015.01.009
Kumar, R., Saha, S., Dhaka, S., Kurade, M. B., Kang, C. U., Baek, S. H., & Jeon, B. H. (2017). Remediation of cyanide-contaminated environments through microbes and plants: A review of current knowledge and future perspectives. Geosystem Engineering, 20(1), 28–40. https://doi.org/10.1080/12269328.2016.1218303
Leys, N. M., Bastiaens, L., Verstraete, W., & Springael, D. (2005). Influence of the carbon/nitrogen/phosphorus ratio on polycyclic aromatic hydrocarbon degradation by Mycobacterium and Sphingomonas in soil. Applied Microbiology and Biotechnology, 66, 726–736. https://doi.org/10.1007/s00253-004-1766-4
Li, M., Li, Y., Fan, X., Qin, Y., He, Y., & Lv, Y. (2019). Draft genome sequence of Rummeliibacillus sp. strain TYF005, a physiologically recalcitrant bacterium with high ethanol and salt tolerance isolated from spoilage vinegar. Microbiology Resource Announcements, 8(31), e00244-19. https://doi.org/10.1128/MRA.00244-19
Luque-Almagro, V. M., Huertas, M. J., Martínez-Luque, M., Moreno-Vivián, C., Roldán, M. D., García-Gil, L. J., & Blasco, R. (2005). Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Applied and Environmental Microbiology, 71(2), 940–947. https://doi.org/10.1128/AEM.71.2.940-947.2005
Maitra, S. (2018). In situ bioremediation—An overview. Research Journal of Life Sciences, Bioinformatics, Pharmaceutical and Chemical Sciences, 4, 576–598. https://doi.org/10.26479/2018.0406.45
Malmir, N., Fard, N. A., Aminzadeh, S., Moghaddassi-Jahromi, Z., & Mekuto, L. (2022). An overview of emerging cyanide bioremediation methods. Processes, 10(9), 1724. https://doi.org/10.3390/pr10091724
Mekuto, L., Jackson, V. A., & Ntwampe, S. K. O. (2013). Biodegradation of free cyanide using bacillus Sp. consortium dominated by bacillus safensis, lichenformis and tequilensis strains: A bioprocess supported solely with whey. Journal of Bioremediation and Biodegradation, S18, 004. https://doi.org/10.4172/2155-6199.S18-004
Murphy, B., Sparacio, T., & Shields, W. (2005). Manufactured gas plants—processes, historical development, and key issues in insurance coverage disputes. Environmental Forensics., 6(2), 161–173. https://doi.org/10.1080/15275920590952847
Neina, D. (2019). The role of soil pH in plant nutrition and soil remediation. Applied and Environmental Soil Science, 1–9. https://doi.org/10.1155/2019/5794869
Njoku, K. L., Akinyede, O. R., & Obidi, O. F. (2020). Microbial remediation of heavy metals contaminated media by Bacillus megaterium and Rhizopus stolonifer. Scientific African, 10, e00545. https://doi.org/10.1016/j.sciaf.2020.e00545
Novak, D., Franke-Whittle, I. H., Pirc, E. T., & Jernan, V. (2013). Biotic and Abiotic process contribute to success anearobic degradation of cyanide by UASB rector biomass treating brewery. Water Research, 47(11), 3644–3653. https://doi.org/10.1016/j.watres.2013.04.027
Nvokoro, O., & Dibua, M. E. U. (2014). Degradation of soil cyanide by single and mixed cultures of Pseudomonas stutzeri and Bacillus subtilis. Archives of Industrial Hygiene and Toxicology, 65(1), 113–119. https://doi.org/10.2478/10004-1254-65-2014-2449
Ouriache, H., Moumed, I., Arrar, J., Namane, A., & Lounici, H. (2020). Influence of C/N/P ratio evolution on biodegradation of petroleum hydrocarbons-contaminated soil. Algerian Journal of Environmental Science and Technology, 6, 1604–1611.
Pandey, S., Ghosh, P. K., Ghosh, S., De, T. K., & Maiti, T. K. (2013). Role of heavy metal resistant Ochrobactrum sp. and Bacillus spp. strains in bioremediation of a rice cultivar and their PGPR like activities. Journal of Microbiology, 51, 11–17. https://doi.org/10.1007/s12275-013-2330-7
Patel, S., & Gupta, R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. International Journal of Systematic and Evolutionary Microbiology, 70(1), 406–438. https://doi.org/10.1099/ijsem.0.003775
Paul, O., Jasu, A., Lahiri, D., Nag, M., & Ray, R. R. (2021). In situ and ex situ bioremediation of heavy metals: The present scenario. Journal of Environmental Engineering and Landscape Management, 29(4), 454–469. https://doi.org/10.3846/jeelm.2021.15447
Razanamahandry, L. C., Karoui, H., Andrianisa, H. A., & Yacouba, H. (2017). Bioremediation of soil and water polluted by cyanide: A review. African Journal of Environmental Science and Technology, 11(6), 272–291. https://doi.org/10.5897/AJEST2016.2264
Sher, S., Sultan, S., & Rehman, A. (2021). Characterization of multiple metal resistant Bacillus licheniformis and its potential use in arsenic contaminated industrial wastewater. Applied Water Science, 11, 1–7. https://doi.org/10.1007/s13201-021-01407-3
Shishir, T. A., Mahbub, N., & Kamal, N. E. (2019). Review on bioremediation: a tool to resurrect the polluted rivers. Pollution, 5(3), 555–568. https://doi.org/10.22059/poll.2019.272339.558
Stout, S., & Brey, A. (2019). Appraisal of coal- and coke-derived wastes in soils near a former manufactured gas plant, Jacksonville, Florida. International Journal of Coal Geology, 213, 103265. https://doi.org/10.1016/j.coal.2019.103265
Sut, M., Fischer, T., Repmann, F., Raab, T., & Dimitrova, T. (2012). Feasibility of field portable near infrared (NIR) spectroscopy to determine cyanide concentrations in soil. Water Air and Soil Pollution, 223(8), 5495–5504. https://doi.org/10.1007/s11270-012-1298-y
Sut, M., Repmann, F., & Raab, T. (2014). Retardation of iron-cyanide complexes in the soil of a former Manufactured Gas Plant site. Journal of Environmental Science and Health, Part A. https://doi.org/10.1080/10934529.2015.981116
Sut-Lohmann, M., & Raab, T. (2017). Quick detection and quantification of iron-cyanide complexes using fourier transform infrared spectroscopy. Environmental Pollution, 227, 64–72. https://doi.org/10.1016/j.envpol.2017.04.052
Sut-Lohmann, M., Boldt-Burisch, K., & Raab, T. (2015). Influence of arbuscular mycorrhizal fungus (AMF) on degradation of iron-cyanide complexes. In European Geosciences Union, General Assembly 2015, Vienna, Austria, 13 April-17 May 2015. European Geophysical Society.
Wehrer, M., Rennert, T., Mansfeldt, T., & Totsche, K. U. (2011). Contaminants at former manufactured gas plants: sources, properties, and processes. Critical Reviews in Environmental Science and Technology, 41, 1883–1969. https://doi.org/10.1080/10643389.2010.481597
Wu, C. F., Xu, X. M., Zhu, Q., Deng, M. C., Feng, L., Peng, J., & Wang, J. H. (2014). An effective method for the detoxification of cyanide-rich wastewater by Bacillus sp. CN-22. Applied Microbiology and Biotechnology, 98, 3801–3807. https://doi.org/10.1007/s00253-013-5433-5
Xiong, C., & Lu, Y. (2022). Microbiomes in agroecosystem: Diversity, function and assembly mechanisms. Environmental Microbiology Reports, 14(6), 833–849. https://doi.org/10.1111/1758-2229.13126
Zarime, N. A., Solemon, B., Wan Yaacob, W. Z., Jamil, H., Che Omar, R., & Oyekanmi, A. A. (2023). Effectiveness of artificially synthesized granitic residual soil-supported nano zero-valent iron (Gr-nZVI) as effective heavy metal contaminant adsorbent. Inorganics, 11(3), 131. https://doi.org/10.3390/inorganics11030131
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chojnacka, A., Sut-Lohmann, M., Jonczak, J. et al. An Evaluation of Long-Term Contaminated Soil from a Manufactured Gas Plant for in Situ Biodegradation Potential and as a Source of Ferrocyanide-Degrading Bacteria. Water Air Soil Pollut 235, 342 (2024). https://doi.org/10.1007/s11270-024-07157-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-024-07157-7