Abstract
Spatially restricted gene flow and resulting spatial genetic structure are generally considered as being the primary controlling factors in the dynamics of biparental inbreeding depression in a wide range of plant species. However, wind-pollinated angiosperm trees have not been studied adequately in this respect. The present study analyses the relationships among parental genetic similarity, outcrossing distances, progeny vigour and mortality in Polylepis australis (Rosaceae), a wind-pollinated treeline species endemic to Argentina. We investigated whether spatial genetic structuring occurs in anthropogenically fragmented P. australis woodlands of the Córdoba Mountains. We also performed a controlled crossing experiment using pollen collected from different distances. Genetic variability (using RAPD-PCR), vigour (N-metabolism capacity) and mortality of the resulting progeny were contrasted with progeny from unmanipulated flowers. We found a continuous decrease in parental genetic similarity with spatial distance among mates and an increase in N-metabolism capacity in the progeny produced from pollen at increasing distances, as well as a very high mortality of seedlings resulting from short-distance crosses. Additionally, our results suggest that there is still fragment connectivity in P. australis through long-distance pollen-mediated gene flow.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Restricted gene dispersal through both seeds and pollen may lead to genetic isolation by distance (Wright 1946; Turner et al. 1982). Under such circumstances, the relatedness between individual plants would decrease with increasing distance between mates (e.g. Wright 1946; Turner et al. 1982; Sokal and Wartenberg 1983). Inbreeding is the crossing between closely related individuals, leading to an increase in the frequency of homozygotes and a decrease of heterozygotes. Crosses between neighbouring plants may lead to biparental inbreeding depression, defined as a reduction in the fitness of the sexual offspring from related parents relative to the sexual offspring from unrelated parents (Levin 1981; van Rossum and Triest 2007).
Spatial clustering of genotypes and biparental inbreeding in natural populations has been demonstrated in various animal-pollinated forest trees (e.g. Garcia Collevatti et al. 2001; Tsumura and Muhammadk 2001; Lemes et al. 2007) and for a number of conifers (e.g. Mitton et al. 1997; Epperson and Chung 2001; Krakowski et al. 2003; Wang et al. 2004). However, studies that properly demonstrate biparental inbreeding depression in tree species are very rare in the available literature (Koptur 1984; Stacy 2001; Hirayama et al. 2005) and even less is known on biparental inbreeding depression in wind-pollinated woody angiosperms.
Inbreeding depression is frequently cited as being a consequence of anthropogenic forest fragmentation in species with a long evolutionary history of large continuous populations (Stacy 2001). Consequently, it is crucial to gather knowledge on reproduction processes and on gene flow in order to conserve their genetic resources (Kundu 1999).
The wind-pollinated woody species of the genus Polylepis are endemic to the South American high mountains. The once extensive forests of these mountains are today mostly restricted to relict stands due to anthropogenic fires, browsing by livestock and deforestation (Ellenberg 1958; Kessler 1995a; Cingolani et al. 2008; Coblentz and Keating 2008). Protection of the high-mountain Polylepis woodlands of South America has been given high priority by the World Conservation Monitoring Center because they belong to one of the most threatened woodland ecosystems in the world (UNEP-WCMC 2004). Polylepis australis, the southernmost Polylepis species, is endemic to the mountains of Argentina. Due to three centuries of human intervention, such as domestic grazing and fires, many regions previously occupied by P. australis woodlands now comprise only small woodland fragments and/or isolated Polylepis trees (Renison et al. 2006; Cingolani et al. 2008), which often show poor recruitment (Renison and Cingolani 1998) and inferior seed viability (Renison et al. 2004).
Decreased seed development might result from either post-zygotic self-incompatibility or early acting biparental inbreeding depression. However, several authors report that effects of biparental inbreeding are more pronounced in fitness components of the seedlings rather than in earlier life stages (e.g. Waser and Price 1994; Trame et al. 1995). In accordance, a previous study on P. australis showed no significant differences in seed germination following hand outcrosses with pollen from varying distances (Seltmann et al. 2008). Thus, we tested if distances between pollen donors and recipient plants influenced progeny vigour (measured as N-metabolism capacity) and mortality (proportion of dead seedlings) by combining pollination experiments with molecular genetic studies on seed and seedling progenies, and measurements of progeny vigour and mortality.
Our objectives were: (1) to examine whether there is an association between genetic similarity and distance between trees, (2) to determine whether outcrossing distances influence genetic variability, N-metabolism capacity and mortality of the progeny in hand pollinations, (3) to compare genetic variability, N-metabolism capacity and mortality of the progeny from hand pollination with progeny from unmanipulated flowers and (4) on the basis of our results, discuss what scenarios of gene flow are most likely to be occurring in the Córdoba Mountains. This is especially important because gene flow is one of the key factors determining species responses to fragmentation (Burczyk et al. 2004).
Materials and methods
Study species and area
The genus Polylepis R. & P. (Rosaceae, Sanguisorbeae), endemic to the South American high mountains, is distributed from south Venezuela to central Argentina and is represented by about 28 species (Simpson 1979; Simpson 1986; Kessler 1995a, 1995b; Schmidt-Lebuhn et al. 2006). The small to large, wind-pollinated trees and shrubs (Simpson 1986) constitute the potential natural vegetation of a large part of the high Andes and other South American mountains up to 5,000 m a.s.l. (Ellenberg 1958; Kessler 1995a; Cingolani et al. 2008).
In the Córdoba mountains of central Argentina (31º 34′ S, 64º 50′ W), stands of P. australis are located between 900 and 2,900 m a.s.l. (Marcora et al. 2008). The main flowering phase occurs between September and October. The anemophilous flowers are arranged in racemiform pendulous inflorescences that are produced annually. Self-pollination is prevented by protogyny. Fruits are single seeded nutlets (mean mass 5 mg) enclosed in a turbinate and winged receptaculum and are dispersed between January and March.
The Córdoba mountains are characterised by a mean annual temperature of 8°C and a mean annual precipitation of 840 mm (Cabido et al. 1987). The present-day vegetation consists of a mosaic of tussock grasslands, grazing pasture, granite outcrops, eroded areas with exposed rock surfaces and P. australis woodlands or scrublands (12% of the surface; Cingolani et al. 2004). Pollination treatments were performed in two woodland fragments within the Yuspe river basin that has been highly impacted by human activities and has an average of 3% woodland and scrubland cover—well below the estimated potential of 52% and average present cover of 12% for the entire Córdoba Mountains (Cingolani et al. 2004, 2008; Renison et al. 2006). The two selected forest fragments were separated by a distance of 1 km (fragment 1: S31°24′09″; W64°4′21″, 1,990 m a.s.l., fragment 2: S31°24′39″; W64°48′28″, 2,220 m a.s.l.). Both fragments were situated in deep ravines along small streams, where impact of fires and livestock was less severe than in the surrounding matrix and forests have virtually disappeared during the past 400 years (Cingolani et al. 2008). Because of the rough topography, tree numbers are hard to estimate, but each fragment comprises a total of >1,000 trees.
Pollination treatments, seedling cultivation and mortality
Seeds of P. australis were obtained by the following pollination treatments that were conducted simultaneously in 20 randomly selected P. australis focal females within the two woodland fragments: (1) no experimental manipulation (open pollination OP). (2) Hand cross-pollination (HCP) with pollen belonging to three distance classes: (i) HCP-1, within a fragment of trees—pollen donors were situated within 20 m of the focal female; (ii) HCP-2, between closely separated fragments—pollen donors were situated 1 km away; (iii) HCP-3, between widely separated fragments—pollen donors were situated 30 km away.
In order to accomplish hand pollination, we collected flowers from 8 to 10 different pollen donors per distance class in the male phase and carefully applied pollen to the receptive stigma using the anthers themselves as pollen applicators. In order to achieve comparable results on genetic diversity of the progeny, attention was paid that the distance between pollen donors was constant across treatments (15–20 m). Except for the baseline OP, flowers were bagged in the budding stage within pollen-proof bags (glassine shoot bags, Seedburo Equipment Company, Chicago). When flowers were receptive females, bags were removed carefully, hand pollinations were performed and inflorescences were subsequently enclosed in the same bag. After fruit maturation, seeds were collected and germinated in a climate chamber with a warm white light source at a temperature of 20/10°C (12 h of light/12 h of darkness). Resulting seedlings were transplanted to individual flower pots of 10 cm diameter immediately after germination and cultivated in the greenhouse under the same conditions for the following 40 days. A biweekly inspection of all seedlings provided survival counts.
Genetic analysis (RAPD-PCR)
In order to analyse genetic similarity between focal females and pollen donors (parent populations), we randomly collected leaf tissue from five focal females of one woodland fragment and from five pollen donors from each of the three respective distance classes −20 m, 1 and 30 km, representing a sample of the same parental individuals used in the outcrossing experiment. Due to the fact that all collected flowers from varying pollen donor trees were treated as one “unit,” it was not possible to assign each female to only one pollen donor. However, based on the assumption that all pollen donors donated pollen to the outcrossing experiment, genetic comparisons were made between all focal females and all potential pollen donors.
We used RAPDs in order to analyse the genetic diversity. In spite of the often raised concern on the reliability of RAPDs, they have been shown to yield similar results as other anonymous marker systems (Nybom 2004). RAPDs provided reliable genetic diversity patterns when applied to P. australis, as evidenced by a previous study where RAPD and ISSR markers gave similar values of diversity (Julio et al. 2008). All seedlings that were not used for the evaluation of N-metabolism capacity (see below) were used as a source of plant material for studies on genetic diversity of the progeny resulting from the different pollination treatments (hereafter referred to as progeny populations). Because of the high mortality of seedlings resulting from pollinations within a fragment (HCP-1), genetic analysis could only be performed on progeny resulting from pollination manipulations between fragments (HCP-2 and HCP-3) and for OP, which resulted in a total sample of 54 seedlings (n = 14 for HCP-2, n = 20 for HCP-3, n = 20 for OP).
Leaf tissue of parent plants and progeny was dried for 24–48 h in silica gel, and genomic DNA was extracted from ~25 mg portions of the dried leaf material following the protocol described by Qiagen (2003). Amplifications of DNA were performed in reaction volumes of 10 μl containing 0.8-μl DNA (10 ng/μl), 0.6 μl of primer (Roth), 1 μl of each dNTP (peqlab), 1 μl 10× buffer (Qbiogene), 0.1 μl Taq Polymerase (5U/μl, Qbiogene) and 6.5 μl H2O. PCR was carried out in an Eppendorf Mastercycler gradient which was programmed for one cycle of 2 min at 94°C followed by 36 cycles of 12 s at 94°C, 45 s at 36°C and 120 s at 72°C with a final cycle of 7 min at 72°C. Separate PCRs were performed for each primer pair. An initial screening of 40 RAPD primers (Roth sets D, N) was performed in order to test amplification profiles for polymorphism, readability and reproducibility. This resulted in the selection of 14 primers for use in the analysis. DNA fragments were separated by electrophoresis in 2% agarose gels with a Tris–acetate–EDTA (TAE) buffer system at 150 V for 150 min and stained with ethidium bromide. DNA bands were then visualised by UV light and documented using a video camera. Gel pictures were analysed with the software RFLPSCAN PLUS Version 3.0 (Scanalytics); all bands between 260 and 2,600 bp were scored.
N-metabolism capacity (15N uptake)
Due to the important role of N-metabolism in biological and ecological systems, 15N stable isotope techniques are used in a broad range of ecological studies (e.g. Hofmann et al. 1997; Jung et al. 1997; 1999; Schulz et al. 2001; Russow et al. 2002). One possible application of stable isotopes like 15N is their use as tracers in detecting the destiny of N-atoms, and on this basis, to determine the N-metabolism capacity of plant material (e.g. Möcker et al. 1998; Jung et al. 1999; Sutter et al. 2002). As plant growth is dependent on an adequate nitrogen supply in order to form amino acids, proteins, nucleic acids and other cellular constituents (López-Cantarero et al. 1997), there is a potentially far-reaching influence of nitrogen metabolism on plant development, and nitrogen limitation may have important consequences for secondary metabolism and plant fitness (Stitt et al. 2002). There is also evidence that effective N-uptake and active N-metabolism may be the crucial factors for survival of tree seedlings at treelines (Weih and Karlsson 1999a, b).
We subsequently applied the 15N tracer technique to specify the N-metabolism capacity of the 6-week-old P. australis progeny resulting from different pollination manipulations using the level of 15N uptake as a parameter of seedling vigour.
Two to three seedlings per maternal tree were chosen (from 6 to 14 maternal trees per pollination treatment), resulting in a total of 84 seedlings. Their roots were washed carefully in distilled water. For the 15N tracer studies, roots were incubated in test tubes with standard media with constant stable isotope abundance (K15NO3 with 95 at.% 15N as a nitrogen source) for 5 h. Thereafter, seedling roots were sectioned and the plant material was frozen at −22°C before 15N sample processing. Each of the 100-mg plant material samples was then digested in 2-ml concentrated H2SO4. The resulting (15NH4)2SO4 solutions were distilled using a modified Micro-Kjeldahl apparatus in accordance with Faust et al. (1981). The nitrogen amount was measured by titration with 0.02 N H2SO4. Thereafter, the sample was reduced down to its pure ammonium salt by boiling. For the emission spectrometric isotope analysis of 15N (15N at.%), the nitrogen as ammonium sulphate was dissolved in distilled water and 25 ml of the solution was used. Isotope abundance was determined by the 15N analyser system (NOI-7; Fischer Analyseninstrumente, Leipzig, Germany), performing five parallel measurements per sample.
Statistical analysis
Genetic analysis
Bands were scored as present or absent. Due to the dominant nature of RAPD markers, we concentrated on band-based analyses to describe genetic similarity. In order to quantify the genetic similarity between focal females and pollen donors for each distance treatment, the proportion of bands shared between two individuals was calculated as twice the number of shared bands divided by the sum of all bands occurring in the two individuals (Dice similarity coefficient), according to Nei and Li (1979). Because the pollen donors were counted as one unit, an average of the Dice similarity coefficients per focal female was calculated. In order to detect differences in genetic similarity resulting from the different outcrossing distances, a one-way ANOVA and Tukey’s HSD test was performed using SPSS 12.0 (SPSS Inc. 2003).
In order to analyse genetic diversity of the progeny populations, again a band-based approach was chosen. We calculated mean Dice dissimilarity among individuals of a given group as a proxy for genetic diversity. We tested differences among groups with a customised Mantel test, where a matrix containing the Dice dissimilarities was tested against a design matrix where group membership and difference were coded with 1 and 0, respectively (9,999 permutations). The genetic variability of each progeny population was also estimated as the proportion of polymorphic loci related to all 135 bands scored, and the average gene diversity over all loci was examined using TFPGA ver.1.3 (Miller 1997). To this end, we had to assume that populations were in Hardy–Weinberg equilibrium. Since the proportion of polymorphic loci is sample-size dependent, we took random sub-samples of n = 14 individuals for OP and HCP-3 to estimate this parameter of genetic variability.
Seedling mortality and N-metabolism capacity
Proportions of seedling mortality were compared with Pearsons chi-square test. Data on N-metabolism capacity data were ln-transformed to achieve a normal distribution of residuals. In order to contrast 15N uptake of the progeny resulting from the different pollination manipulations, a Randomized Block ANOVA with Scheffé test was performed regarding ‘pollination treatment’ as the fixed factor and ‘tree’ as the block factor.
For seedling mortality, the magnitude of inbreeding depression was calculated as the difference between mortality of seedlings from within-fragment crosses and the mortality of seedlings from between-fragment crosses.
For N-metabolism capacity, we calculated the severity of inbreeding depression as follows:
-
1—(15N uptake of 20 m crosses)/(15N uptake of 30 km crosses) and
-
1—(15N uptake of 1 km crosses)/(15N uptake of 30 km crosses) (modified following Barrett and Kohn 1991).
Statistical analyses were performed using SPSS 12.0 (SPSS Inc. 2003) and NCSS 2001 (Hintze 2001) software.
Results
Genetic analysis
The 14 primers used in the RAPD analysis of leaf tissue of 20 parent trees and of 54 seedlings of the progeny resulted in 135 polymorphic scorable bands with each plant being distinguishable as a single RAPD phenotype.
Genetic similarity between focal females and pollen donors
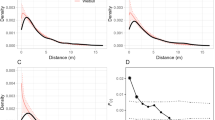
Genetic similarity was highest in parental trees located within a woodland fragment (HCP-1, mean Dice similarity: 0.727 ± 0.025 SD; Fig. 1) and significantly higher than those of parental trees separated by a distance of 30 km (one-way ANOVA; df = 14; P < 0.05). The values of genetic similarity were very similar for parental trees separated by a distance of 1 and 30 km (HCP-2, mean: 0.699 ± 0.014; HCP-3, mean: 0.689 ± 0.023; Fig. 1).
Genetic similarity (proportion of bands shared, Dice similarity coefficient) of focal females and pollen donors of Polylepis australis from three different distance classes (HCP-1: 20 m; HCP-2: 1 km; HCP-3: 30 km; n = 5 for focal females and five pollen donors for each distance). The values plotted are based on the mean genetic similarity of a focal female to its five pollen donors. Different letters indicate significant differences (Tukey’s HSD test, P < 0.05)
Genetic diversity of the progeny populations
All measures of genetic diversity, i.e. the mean Dice dissimilarity, the proportion of polymorphic loci and the average gene diversity reached highest values in the progeny originating from open-pollinated flowers (OP; Table 1). Considering the progeny populations of hand-outcrossed flowers, the proportion of polymorphic loci and the average gene diversity were slightly larger if pollen came from the more distant donors. However, this was not the case for Dice dissimilarity (Table 1). A Mantel test based on the Dice dissimilarity values revealed that samples within groups were significantly more similar than samples belonging to different groups; the standardised Mantel statistic (akin to a correlation coefficient) was r M = 0.20 (P < 0.0001).
Seedling mortality and N-metabolism capacity
Mortality was clearly highest in seedlings resulting from outcrossings within a fragment (HCP-1: 86.3%) and lowest in seedlings from outcrossings between fragments (HCP-2: 63.9%; HCP-3: 66.4%), while mortality was intermediate in seedlings resulting from OP (76.6%) (Chi-squared test, χ2 = 18.1; df = 3; P < 0.001).
The pollination treatment (i.e. the outcrossing distance) had a significant effect on the amount of 15N that was incorporated by the resulting progeny (Randomized Block ANOVA; df = 3; P < 0.01), while there was no effect of the block factor ‘tree’ (df = 16; P = 0.83). The 15N uptake of the progeny increased with outcrossing distance (Fig. 2). We did not find any significant differences between OP and outcrossings between widely separated fragments (HCP-3), whereas seedlings resulting from outcrossings within a fragment (HCP-1) and from outcrossings between closely separated fragments (HCP-2) incorporated significantly less 15N.
N-metabolism capacity of Polylepis australis as measured by 15N uptake from seedlings resulting from open pollination (OP) and from three different distance classes (HCP-1: 20 m; HCP-2: 1 km; HCP-3: 30 km). The values plotted are based on the mean 15N uptake of the progeny of each maternal tree (two to three seedlings per maternal tree; OP: n = 8 trees; HCP-1: n = 6 trees; HCP-2: n = 14 trees; HCP-3: n = 13 trees). Different letters indicate significant differences (Tukey’s HSD test, P < 0.05)
In case of seedling mortality, the magnitude of inbreeding depression for within-fragment crosses was 20% compared to long-distance crosses and 22% compared to crosses between closely separated fragments. In case of N-metabolism capacity, inbreeding depression compared to long-distance crosses was 14.1% for within-fragment crosses and 9.6% for crosses between closely separated fragments.
Discussion
Our study provides evidence of a spatial genetic structuring in P. australis, an increase in progeny nitrogen metabolism capacity with increasing outcrossing distances, and a high mortality in seedlings resulting from hand crosses within a fragment. These findings indicate significant biparental inbreeding depression, especially in plants that are crossed with nearby neighbours. Genetic similarity between focal females and pollen donors may be regarded as the underlying factor for the biparental inbreeding depression revealed in this study. However, as seedlings of long-distance crosses (30 km) showed high fitness and low mortality despite relatively high levels of genetic similarity of mates, biparental inbreeding depression in P. australis should become apparent only at very high levels of genetic similarity.
Our findings are in line with Trame et al. (1995) who reported a negative relationship between genetic similarity and spatial distance of parent plants as well as a significant biparental inbreeding depression for Agave schottii (Agavaceae). However, to our knowledge there are as yet no studies that have tested this relationship directly using genetic differences and progeny vigour or which have clearly detected evidence of the occurrence of biparental inbreeding depression in wind-pollinated angiosperm trees.
As Seltmann et al. (2008) did not find any difference in seed germination between crosses of nearby and distant fragments, we conclude that biparental inbreeding depression bears more significance to progeny fitness and mortality than to germination; a finding that is in line with the assumptions of several authors like Waser and Price (1994), Trame et al. (1995) and Hardner et al. (1998). Correspondingly, coefficients of inbreeding depression were clearly higher in within-fragment crosses than in crosses between nearby fragments, with the former resulting in a very high mortality of seedlings. Thus, mating between relatives may lead to inbred progeny, which has reduced chances to become established in the population.
However, results from the un-manipulated OP treatment and the high genetic similarity observed even for individuals 30 km apart indicate that there is still long-distance pollen flow buffering inbreeding depression under current levels of fragmentation. This assumption is supported by the finding of low genetic differentiation of P. australis in the Córdoba mountains (0.001 ≤ ϕst ≤ 0.05; Julio et al. 2008) and of other species of the genus (Schmidt-Lebuhn et al. 2006). In accordance, Seltmann et al. (unpubl.) found pollen dispersal distances of P. australis ranging up to 80 km from the nearest population and documented an exceptionally long period of pollen viability (47% of viable pollen grains after 128 h following pollen release).
The fact that all estimates of genetic variability were highest in seedlings resulting from OP may be explained by the scenario that our open pollinated flowers may still have received long distant as well as short-distant pollen, and from many more than 10 donors.
It is suggested that population connectivity through pollen-mediated gene flow may have substantial effects on the persistence of isolated fragments, as gene flow into patches may serve to mitigate the effects of inbreeding depression (Richards 2000). The large fecundity of P. australis and the possibility of (even rare) long-distance pollen flow events should ensure the production of a large enough number of lowly inbred individuals. However, at the same time inbreeding depression provides potential opportunities for purging deleterious alleles and the resultant depression in fitness in future generations (e. g. Wang 2000; Ward et al. 2005).
Nevertheless, we found that effective pollen movement over substantial distances is essential for maintaining seedling fitness. This is reflected especially by the finding of high mortality of seedlings that were fathered by individuals of the same woodland. In consequence, for the natural regeneration of P. australis woodlands, even further increased fragmentation of the remaining stands interrupting pollen exchange due to ongoing land-use practices would involve a potential for increased biparental inbreeding and thus, decreased progeny fitness and increased seedling mortality.
Pollen-mediated gene flow may act to rescue populations that are within the range of pollen dispersal but may have no effect on completely isolated populations (Richards 2000). In accordance, recent research on several conifers supports that the rate of inbreeding was higher in small, disjunct low-density populations (mass-action assumption; Rajora et al. 2000, 2002; Sorensen 2001). We conclude therefore that conservation of the remaining P. australis woodlands in Argentina should be aimed at avoiding any complete isolation of fragments.
Spatial genetic structuring in populations of wind-pollinated trees has been implicated in many conifer populations (e.g. Epperson 1992, 2000; Epperson and Chung 2001), and has been explained by restricted seed and pollen dispersal. The finding of higher genetic similarity of P. australis individuals within a fragment and of near-situated fragments indicates that clustering of related genotypes in P. australis is not only caused by incomplete pollen flow, but also by limited migration distance of seeds. P. australis has gravity-dispersed and wind-dispersed seeds that were never trapped more than 6 m away from seeder trees, and seedlings were never found more than 10 m away (Torres et al. 2008). However, seed dispersal seemed to be more effective than expected, as inbreeding depression occurred not only within, but also among near-distant woodland fragments. As these are located in the same river basin, we presume that seeds may be transported over substantial distances by water, leading to the occurrence of half-sibs in different nearby-woodland fragments. Nevertheless, even moderate limits to dispersal are known to cause spatial structuring of genotypes and hence subsequent biparental inbreeding (Epperson and Chung 2001).
References
Barrett CH, Kohn JR (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, Oxford
Burczyk J, DiFazio SP, Adams WT (2004) Gene flow in forest trees: how far do genes really travel? For Gen 11:1–14. doi:10.1080/10623320490432434
Cabido M, Breimer R, Vega G (1987) Plant communities and associated soil types in a high plateau of the Córdoba mountains, central Argentina. Mt Res Dev 7:25–42. doi:10.2307/3673322
Cingolani AM, Renison D, Zak MR, Cabido M (2004) Mapping vegetation in a heterogeneous mountain rangeland using landsat data: an alternative method to define and classify land-cover units. Remote Sens Environ 92:84–97. doi:10.1016/j.rse.2004.05.008
Cingolani A, Renison D, Tecco P, Gurvich D, Cabido M (2008) Predicting cover types in a mountain range with long evolutionary grazing history: a GIS approach. J Biogeogr 35:538–551. doi:10.1111/j.1365-2699.2007.01807.x
Coblentz D, Keating PL (2008) Topographic controls on the distribution of tree islands in the high Andes of south-western Ecuador. J Biogeogr 35:2026–2038. doi:10.1111/j.1365-2699.2008.01956.x
Ellenberg H (1958) Wald oder Steppe? Die natürliche Pflanzendecke der Anden Perus. Umschau 22:645–681
Epperson BK (1992) Spatial structure of genetic variation within populations of forest trees. New For 6:257–278. doi:10.1007/BF00120648
Epperson BK (2000) Spatial genetic structure and non-equilibrium demographics within plant populations. Plant Species Biol 15:269–279. doi:10.1046/j.1442-1984.2000.00046.x
Epperson BK, Chung MG (2001) Spatial genetic structure of allozyme polymorphisms within populations of Pinus strobus (Pinaceae). Am J Bot 88:1006–1010. doi:10.2307/2657081
Faust H, Bornhak H, Hirschberg K, Jung K, Junghans P, Krumbiegel P (1981) 15N-Anwendung in der Biochemie, Landwirtschaft und Medizin. Schriftenr Anwendung Isotopen Kernstrahlungen Wiss Tech 5:1–93
Garcia Collevatti R, Grattapaglia D, Hay JD (2001) High resolution microsatellite based analysis of the mating system allows the detection of significant biparental inbreeding in Caryocar brasiliense, an endangered tropical tree species. Heredity 86:60–67. doi:10.1046/j.1365-2540.2001.00801.x
Hardner CM, Potts BM, Gore PL (1998) The relationship between cross success and spatial proximity of Eucalyptus globulus ssp. globulus parents. Evolution 52:614–618. doi:10.2307/2411096
Hintze JL (2001) NCSS 2001. Number cruncher statistical systems. Kayville, Utah
Hirayama K, Ishida K, Tomaru N (2005) Effects of pollen shortage and self-pollination on seed production of endangered tree, Magnolia stellata. Ann Bot 95:1009–1015. doi:10.1093/aob/mci107
Hofmann D, Jung K, Bender J, Gehre M, Schüürmann G (1997) Using natural isotope variations of nitrogen in plants as an early indicator of air pollution stress. J Mass Spectrom 32:855–863. doi:10.1002/(SICI)1096-9888(199708)32:8<855::AID-JMS544>3.0.CO;2-J
Julio N, Sobral A, Rondan Dueñas J, Di Rienzo J, Renison D, Hensen I (2008) RAPD and ISSR markers indicate diminished gene flow due to recent fragmentation of Polylepis australis woodlands in central Argentina. Biochem Syst Ecol 36:329–335. doi:10.1016/j.bse.2007.10.007
Jung K, Gebauer G, Gehre M, Hofmann D, Weißflog L, Schüürmann G (1997) Anthropogenic impacts on natural nitrogen isotope variations in Pinus sylvestris stands in an industrially polluted area. Environ Pollut 97:175–181. doi:10.1016/S0269-7491(97)00053-5
Jung K, Kaletta K, Segner H, Schüürmann G (1999) 15N metabolic test for the determination of phytotoxic effects of chemical and contaminated environmental samples. Environ Sci Pollut Res 6:72–76. doi:10.1007/BF02987551
Kessler M (1995a) Polylepis-Wälder Boliviens: Taxa, Ökologie, Verbreitung und Geschichte. Diss Bot 246:1–303
Kessler M (1995b) Revalidación de Polylepis rugulosa Bitter (Rosaceae). Gayana Bot 52:49–51
Koptur S (1984) Outcrossing and pollinator limitation of fruit set: breeding systems of neotropical Inga trees (Fabaceae: Mimosoideae). Evolution 38:1130–1143. doi:10.2307/2408445
Krakowski J, Aitken SN, El-Kassaby YA (2003) Inbreeding and conservation genetics in whitebark pine. Conserv Genet 4:581–593. doi:10.1023/A:1025667700479
Kundu SK (1999) The mating system and genetic significance of polycarpy in the neem tree (Azadirachta indica). Theor Appl Genet 99:1216–1220. doi:10.1007/s001220051327
Lemes MR, Grattapaglia D, Grogan J, Proctor J, Gribel R (2007) Flexible mating system in a logged population of Swietenia macrophylla King (Meliaceae): implications for the management of a threatened neotropical tree species. Plant Ecol 192:169–179. doi:10.1007/s11258-007-9322-9
Levin DA (1981) Dispersal versus gene flow in plants. Ann Miss Bot Garden 68:233–253. doi:10.2307/2398797
López-Cantarero I, Ruiz JM, Hernandez J, Romero L (1997) Nitrogen metabolism and yield response to increases in nitrogen-phosphorus fertilization: improvement in greenhouse cultivation of eggplant (Solanum melongena Cv. Bonica). J Agric Food Chem 45:4227–4231. doi:10.1021/jf970213g
Marcora P, Hensen I, Renison D, Seltmann P, Wesche K (2008) The performance of Polylepis australis trees along their entire altitudinal range: implications of climate change for their conservation. Div Distr 14:630–636. doi:10.1111/j.1472-4642.2007.00455.x
Miller MP (1997) Tools for population genetic analysis (TFPGA), a windows program for the analysis of allozyme and molecular population genetic data. Version 1.3. Department of Biological Sciences, Northern Arizona University, USA, Flagstaff
Mitton JB, Latta RG, Rehfeld GE (1997) The pattern of inbreeding in Washoe Pine and survival of inbred progeny under optimal environmental conditions. Silvae Genet 46:215–219
Möcker D, Jung K, Förstel H, Schüürmann G (1998) Isotopic and enzymatic investigations into the assimitation and effect of NO2 on C3 and C4 plants. J Appl Bot 72:186–190
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273. doi:10.1073/pnas.76.10.5269
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155. doi:10.1111/j.1365-294X.2004.02141.x
Qiagen (2003) DNeasy Plant Mini Kit and DNeasy Plant Maxi Kit. In: Handbook 15, Qiagen Distributors
Rajora OP, Mosseler A, Major JE (2000) Indicators of population viability in red spruce, Picea rubens. II. Genetic diversity, population structure, and mating behavior. Can J Bot 78:941–956. doi:10.1139/cjb-78-7-941
Rajora OP, Mosseler A, Major JE (2002) Mating system and reproductive fitness traits of eastern white pine (Pinus strobus) in large, central versus small, marginal populations. Can J Bot 80:1173–1184. doi:10.1139/b02-105
Renison D, Cingolani AM (1998) Experiencias en germinación y reproducción vegetativa aplicados a la reforestación con Polylepis australis (Rosaceae) en las Sierras Grandes de Córdoba, Argentina. Agriscientia 15:47–53
Renison D, Hensen I, Cingolani AM (2004) Anthropogenic soil degradation affects seed viability in Polylepis australis mountain forests of central Argentina. For Ecol Man 196:327–333
Renison D, Hensen I, Suarez R, Cingolani AM (2006) Cover and growth habit of Polylepis woodlands and shrublands in the mountains of central Argentina: human or environmental influence? J Biogeogr 33:876–887. doi:10.1111/j.1365-2699.2006.01455.x
Richards CM (2000) Inbreeding depression and genetic rescue in a plant metapopulation. Am Nat 155:383–394. doi:10.1086/303324
Russow R, Kupka HJ, Gotz A, Apelt B (2002) A new approach to determining the content and 15N abundance of total dissolved nitrogen in aqueous samples: TOC analyser-QMS coupling. Isotopes Environ Health Stud 38:215–225. doi:10.1080/10256010208033267
Schmidt-Lebuhn AN, Kumar M, Kessler M (2006) An assessment of the genetic population structure of two species of Polylepis Ruiz & Pav. (Rosaceae) in the Chilean Andes. Flora 201:317–325
Schulz H, Gehre M, Hofmann D, Jung K (2001) Nitrogen isotope ratios in pine bark as an indicator of N emissions from anthropogenic sources. Environ Monit Assess 69:283–297. doi:10.1023/A:1010705907525
Seltmann P, Cocucci AA, Renison D, Cierjacks A, Hensen I (2008) Mating system, outcrossing distance effects and pollen availability in the wind-pollinated treeline species Polylepis australis (Rosaceae). Basic Appl Ecol 10:52–60
Simpson BB (1979) A revision of the Genus Polylepis (Rosaceae: Sanguisorbeae). Smithsonian Institution Press, Washington
Simpson BB (1986) Speciation and specialization of Polylepis in the Andes. In: Vuilleumier F, Monasterio M (eds) High altitude tropical biogeography. Oxford University Press, Oxford, pp 304–316
Sokal RR, Wartenberg DE (1983) A test of spatial autocorrelation analysis using an isolation by distance model. Genetics 105:219–237
Sorensen FC (2001) Effect of population outcrossing rate on inbreeding depression in Pinus contorta var. murrayana seedlings. Scand J For Res 16:391–403. doi:10.1080/02827580152632784
SPSS Inc (2003) SPSS for Windows 12. 0G. SPSS Inc, Chicago
Stacy EA (2001) Cross-fertility in two tropical tree species: evidence of inbreeding depression within populations and genetic divergence among populations. Am J Bot 88:1041–1051. doi:10.2307/2657086
Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible W-R, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53:959–970. doi:10.1093/jexbot/53.370.959
Sutter K, Jung K, Krauss G-J (2002) Effects of heavy metals on the nitrogen on the nitrogen metabolism of the aquatic moss Fontinalis antipyretica L. ex Hedw. Environ Sci Pollut Res 9:417–421. doi:10.1007/BF02987592
Torres RC, Renison D, Hensen I, Suarez R, Enrico L (2008) Polylepis australis’ regeneration niche in relation to seed dispersal, site characteristics and livestock density. For Ecol Man 254:255–260
Trame AM, Coddington AJ, Paige KN (1995) Field and genetic studies testing optimal outcrossing in Agave schottii, a long-lived clonal plant. Oecologia 140:93–100. doi:10.1007/BF00365567
Tsumura Y, Muhammadk N (2001) Microsatellite analysis of the breeding system and seed dispersal in Shorea leprosula (Dipterocarpaceae). Int J Plant Sci 162:155–159. doi:10.1086/317902
Turner ME, Stephens JC, Anderson WW (1982) Homozygosity and patch structure in plant populations as a result of nearest neighbor pollinations. Proc Natl Acad Sci USA 79:203–207. doi:10.1073/pnas.79.1.203
UNEP-WCMC (2004) United Nations Environment Programme, World Conservation Moitoring Center. www.unep-wcmc.org
van Rossum F, Triest L (2007) Fine-scale spatial genetic structure of the distylous Primula veris in fragmented habitats. Plant Biol 9:374–382. doi:10.1055/s-2006-924632
Wang J (2000) Effects of population structures and selection strategies on the purging of inbreeding depression due to deleterious mutations. Genet Res 76:75–86. doi:10.1017/S0016672399004450
Wang T, Aitken SN, Woods JH, Polsson K, Magnussen S (2004) Effects of inbreeding on coastal Douglas fir growth and yield in operational plantations: a model-based approach. Theor Appl Genet 108:1162–1171. doi:10.1007/s00122-003-1534-3
Ward M, Dick CW, Gribel R, Lowe AJ (2005) To self or not to self… A review of outcrossing and pollen-mediated gene flow in neotropical trees. Heredity 95:246–254
Waser NM, Price MV (1994) Crossing distance effects in Delphinium nelsoni: outbreeding and inbreeding depression in progeny fitness. Evolution 48:842–852. doi:10.2307/2410491
Weih M, Karlsson PS (1999a) Growth response of altitudinal ecotypes of mountain birch to temperature and fertilisation. Oecologia 119:16–23. doi:10.1007/s004420050756
Weih M, Karlsson PS (1999b) The nitrogen economy of mountain birch seedlings: implications for winter survival. J Ecol 87:211–219. doi:10.1046/j.1365-2745.1999.00340.x
Wright S (1946) Isolation by distance under diverse systems of mating. Genetics 31:39–59
Acknowledgements
The authors thank the Volkswagen foundation for supporting the project; the National Parks Administration (Argentina) for providing working permits; David Smaldone, Cesar García and Ricardo Suarez for assistance in the field; Birgit Mueller and Helga Schroeter for technical support and Joe King for the revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Seltmann, P., Hensen, I., Renison, D. et al. Biparental inbreeding depression, genetic relatedness and progeny vigour in a wind-pollinated treeline species in Argentina. Plant Ecol 205, 155–164 (2009). https://doi.org/10.1007/s11258-009-9605-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11258-009-9605-4