Abstract
Potato is an important part of the traditional Norwegian diet, and the crop faces several challenges with respect to pests and diseases, as well as the increasingly challenging changes in climate. Genome editing may provide tools to improve the resilience of Norwegian potato cultivars to new climate challenges. We have altered the skin colour of two potato cultivars, ‘Desirée’ and ‘Nansen’ from red to yellow, as a proof-of-concept for the use of CRISPR/Cas9 in a Norwegian cultivar. Our method has involved the use of protoplasts and we have grown the regenerants for three successive clonal tuber generations to evaluate the stability of the edited plants over time and under varying temperature conditions in contained rooms in a greenhouse. We found that the protoplast method is well suited to achieving CRISPR/Cas9 applications. The results show that the yellow skin is consistent over the three generations of tuber propagation. We found some suspected somaclonal variation in the protoplast regenerants. Some of the variation which we observed under high temperatures (up to nearly 40ºC) during the second growth cycle, disappeared when cultivated under lower temperatures in the third cultivation cycle.
Key message
Protoplast derived potato obtained RNA knock-outs by CRISPR of F3H in a ribonucleoprotein complex to interrupt with the anthocyanin biosynthesis. The analysis of phenotypes revealed knock-outs in 1–4 alleles. We followed three successive tuber generations, and revealed change of skin colour and how temperature influenced the tuber appearances, as well as some somaclonal variation. Without trials in three tuber generations, we would not have picked up on the influence of temperature on the tubers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is one of the most important clonally propagated crops, being the third most important food crop globally. Global production is estimated to be 470 million metric tons for 2022 (FAOSTAT 2023). Potatoes are relatively easy to cultivate, adaptable to various climates, and can yield a significant amount of food per unit of land. They are a good source of carbohydrates, vitamins, and minerals, making them a valuable staple in many diets around the world (Devaux et al. 2014; Flaten and Hisano 2007). Establishing sustainable, global food security is increasingly important with the effects of climate change become more apparent. The world experiences extreme heat, cold spells at unexpected times of the year, drought, and flooding, as have previously been predicted (Misra 2014; Wheeler and Von Braun 2013). Altered environmental factors may also cause pathogens to evolve and spread to new areas (Singh et al. 2023).
These challenges require swift adaptations in crops and for farmers. This is especially challenging for clonally propagated, polyploid crops, such as potato (Gemenet and Khan 2017). Potato breeding can be very challenging since the cultivated potato is highly heterozygous and also tetraploid. Using conventional crosses will result in rearrangements of the four alleles and end up far from the desired genotype. The improved genotypes may become popular varieties, once obtained, and can last for more than a century. ‘Russet Burbank’, the world’s foremost French fry processing cultivar, originates from a mutation of ‘Burbank’ in 1902 (Bethke et al. 2014). Since potatoes are clonally propagated and preferences vary around the world, there are more than 4000 potato cultivars in the world, and more than 500 on the list in the UK alone (Ghimire 2022). This goes to show that once a variety has caught on, breeding a new one to replace it is hard. To further make breeding complicated, the tetraploidy and the heterozygosity often results in a high likelihood of inbreeding depression (Slater et al. 2014). Twelve to twenty years is the minimum time required for the development and release of a new potato variety (Bonierbale et al. 2020). The tetraploid nature of most cultivated potatoes makes it difficult to breed for traits where all four alleles need to be of the optimal version of the gene(s), such as resistance to diseases. Once a cross has been made, all traits are in play and will be recombined to create novel potato types, but not necessarily with the combination of desired traits. To create an improved variety, a plant breeder must consider many traits simultaneously to create a valuable variety, such as disease resistance, yield, early harvest, starch content and other quality traits (Jansky and Spooner 2018). An increase in the number of breeding goals will exponentially increase the breeding complexity, which usually also increases the time to reach the desired combination of traits. The complexity of breeding combined with a need for quicker adaptation to alter biotic and abiotic aspects are the main factors driving the necessity of innovation, and faster, more precise, or novel alternatives to traditional plant breeding approaches.
Precision breeding such as CRISPR/Cas can be used to accelerate breeding and directly improve specific traits by editing single genes, without altering other parts of the genome. However, most gene technology methods, including CRISPR, utilize genotype specific cell- and tissue culture to make changes to the plant cell. As a result, most research is carried out on model genotypes amenable to cell- and tissue culture regeneration, rather than genotypes of commercial value. Hence, there is still a need for adapting novel breeding technologies (NBT) to commercially interesting varieties.
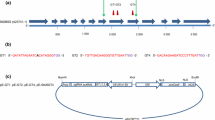
In this study, the CRISPR/Cas9 system is used to knock out flavanone 3-hydroxylase (F3H), a key enzyme in the anthocyanin pathway, in two different potato cultivars: ‘Nansen’, a Norwegian cultivar from 2018 (Potet 2021), and the “model” potato cultivar ‘Desirée’. We hypothesise that knocking out this gene will prevent the formation of the anthocyanidins (pelargonidin, cyanidin and delphinidin in Fig. 1) responsible for red and blue/purple colours in potato (Mishra et al. 2020). As a result, it is likely that both flower and tuber skin colour will change from pink and red to white and yellow, providing a trait which is easy to observe and evaluate in cultivation. This will be a proof-of-concept for using CRISPR for potato research and breeding. In addition, the food industry preparing various potato products for consumption, regards the red skinned varieties of lesser value than yellow skin, since the red skin remain in residues around the yes when peeled. This makes the products less appealing.
Overview of the anthocyanin biosynthesis pathway. The enzyme flavanone 3-hydroxylase (F3H) is essential for formation of the pigments pelargonidin, cyanidin and delphinidin. We hypothesise that knocking out F3H will prevent formation of red and blue colour in the potato tuber.
Modified from Holton and Tanaka (1994)
Methods
Plant material
Solanum tuberosum ‘Nansen’ in vitro plants were a gift from the Norwegian Institute of Bioeconomy Research (NIBIO), Ås, Norway. Solanum tuberosum ‘Desirée’ originated from the Swedish University of Agricultural Sciences (SLU)’s collection in Alnarp, Sweden. Both cultivars were maintained in vitro according to Andersson et al. (2018).
Design of guide RNAs/knockout (KO)
The Solanum tuberosum F3H gene was investigated in Ensembl plants (Cunningham et al. 2021), where a single transcript of the gene was described. This sequence was run through BLASTX and nucleotide BLAST (Altschul et al. 1990) to investigate if the nucleotide sequence is conserved across different potato cultivars. In addition, the first exon of the gene was amplified and sequenced in the Norwegian potato cultivar ‘Nansen’. Genomic DNA was extracted using Qiagen DNeasy plant mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions before PCR amplifying a 486 bp region (primers StF3Hf1 and StF3Hr1 (Supplementary material 1). The PCR product was purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and Sanger sequenced (GATC Biotech, Konstanz, Germany) using StF3Hf1 primer.
Four preliminary guide RNAs (gRNAs, G1-G4) targeting the first exon of F3H were manually selected. The selected guide sequences were subsequently studied in genome sequence data from ‘Desirée’ previously generated (Zhao et al. 2021), to confirm that no regions with allelic variation were selected for the gene editing experiments to ensure that all four alleles were targeted, and to try to avoid similar sequences elsewhere in the genome. Three of the four studied target regions were also conserved in ‘Desirée’, and two of them, G2 and G4, were selected for further experiments, G2 5’-TGAAGAAGAACGTCCAAAAG-3’ and G4 5’-ATGTCTGGTGGCAAGAAAGG-3’.
The guide sequences and the structure of the gene with all exons and the target position in exon 1 is shown in Supplementary Material 2.
Protoplast ribonucleoprotein transfections
Mutations were induced in ‘Nansen’ and ‘Desirée’ at two target sites in F3H, G2 and G4 respectively, by ribonucleoprotein (RNP) transfection of protoplasts. Protoplast isolation and regeneration was performed as previously described, and a detailed protocol can be found in Nicolia et al. 2021 and in (Andersson et al. 2018). Transfection was made using freshly prepared RNPs. For each experiment, 5 µg Cas9 (Thermo Fisher Scientific, Waltham, MA, USA) was preassembled with 0.1 nmol of sgRNA (Synthego, Redwood City, CA, USA) in a total volume of 10 ul, following a 15 min incubation at room temperature. 100.000 protoplasts in 100 ul were added to the preassembled RNPs together with 110 ul of 25% PEG for transfection of ‘Nansen’ and 40% PEG for transfection of ‘Desirée’. The RNP-‘Nansen’ protoplast solution was incubated for 3 min while the RNP-‘Desirée’ protoplast solution was incubated for 30 min, to obtain optimal transfections for the two individual varieties, where 40% PEG for 30 min is a standard protocol for ‘Desirée’ while more gentle conditions with 25% PEG and 3 min incubation was used for the previously untested variety ‘Nansen’. Care was taken to only select one shoot from each callus. Screening of regenerated shoots were made using high-resolution fragment analysis (HRFA), which includes PCR amplification of leaf tissue from 2 to 4 weeks shoots using primers flanking the target regions (see Supplementary Material 3), and indels detected on a 3500 Genetic analyser (Thermo Fisher Scientific, Waltham, USA). Detailed protocols of the HRFA method are described in Andersson et al. 2017 The results from the HRFA screening were used to divide the lines into categories based on the observed mutations; Full KO, where all four alleles were mutated, partial KO, where 1–3 alleles were mutated, and unedited lines, where no mutations were observed in any of the alleles.
Greenhouse cultivation and screening
We chose to do the screening of the various mutants and controls in a greenhouse under Contained Use facilities in the Centre for Plant Research in Controlled Climate (SKP) in NMBU. The temperatures and humidity of the three trials are given in Fig. 2.
A total of 81 regenerated lines were selected for greenhouse cultivation, to investigate observable differences in phenotype based on the number of KO alleles.
In vitro plants were transplanted into commercial potting mix (Tjerbo Gartnerjord, peat: sand: clay, 86:10:4, vol%) in plug trays, one pot for each plant (line) (10–12/05/2021). The plants were acclimatised under a plastic tent in the greenhouse, transferred to 12 cm pots after five weeks (18/6/2021), when a good root system had developed, and further to 12 L potato pots four weeks later (14/7/2021). The plants were grown for 23 weeks in total, before harvesting the tubers. The foliage was removed a couple of weeks before harvest to promote tuber growth and ripening. Upon harvesting, the lines were photographed, and the number of red/yellow tubers was registered for each line.
A second greenhouse clonal tuber generation was performed with the tubers from the first round for all full KO ‘Nansen’ lines (mutation in all four alleles) which gave a yellow phenotype in year one (three lines for G2 and five lines for G4), in addition to three lines for ‘Desirée’ for each guide RNA used. One putatively unedited plant with red skin colour in year 1 was used as a non-edited individual for comparison.
For the second-year cultivation, three tubers were placed directly into peat in 12 L potato pots. Three pots were used for each line (biological replicates), to investigate potential phenotypic differences between clones. The plants were grown for 19 weeks prior to harvesting.
The T3 mutants from protoplast edited potato tubers (originating from the second clonal tuber generation) were planted directly into 12 L potato pots, one tuber in each pot to distinguish between the individual potatoes. This trial was performed from January-April when temperatures fluctuated between 15–25ºC, compared to 15–40 ºC in the greenhouses in summer (Fig. 2).
The duration of the tuber producing clonal tuber generations in contained greenhouse compartments were as follows: first clonal tuber generation (T1) in 2021 23 weeks, second clonal tuber generation (T2) in 2022 19 weeks and the third clonal tuber generation (T3) in 2023 was only 11 weeks, since we considered the colour and phenotype of the tubers to be more important than the yield.
Genotyping putatively edited lines
Genomic DNA was extracted from leaf tissue of selected in vitro lines using Qiagen DNeasy plant mini kit, following the manufacturer’s instructions (Qiagen, Hilden, Germany). The first exon of F3H was amplified using 0.5 µl of genomic DNA, 0.25 µM of each primer (StF3HF1 and StF3HR1, Supplementary material 1) and Phusion polymerase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The annealing temperature was set to 60 °C. The PCR product was cloned into CloneJet ™ pJET 1.2/Blunt and transformed into E. coli DH10B. Twelve colonies were randomly selected for each line and Sanger sequenced by Eurofins Genomics using the pJet 1.2 forward primer (Supplementary Material 4).
Results
The StF3H gene (ID PGSC0003DMG400009715) in the anthocyanin pathway (Fig. 1) was selected as a candidate to visually investigate the efficiency of CRISPR/Cas9 mediated KO in two red skin potato cultivars, the Norwegian ‘Nansen’ and the Dutch ‘Desirée’, to obtain yellow tubers. We also wanted to investigate if there is an observable, additive effect of knocking out all four alleles compared to partial KOs. The analysed F3H region was highly conserved among different cultivars and therefore considered suitable for CRISPR mediated KO.
Primary screening of mutated plants
Regenerated potato lines were screened using high-resolution fragment analysis (HRFA) to investigate the number of mutated alleles (Supplementary Material 5). The lines were labelled according to the preliminary HRFA screening as lines without observed mutations in the target region, with partial KO (one, two or three alleles mutated) or with full KO (all four alleles mutated). The full KO lines were divided into two subcategories, as described for rice by Wu et al. (2020): (1) out of frame mutations in all alleles, where the mutations would lead to a disruption of the reading frame, and (2) in-frame mutations where at least one of the alleles had a mutation that would not disrupt the reading frame (e.g. deletion of 3, 6 or 9 base pairs).
Of the ‘Nansen’ lines edited with G2 and G4, around 7% and 16% respectively, exhibited mutations in all four alleles, whereas 44% (G2) and 61% (G4) of the ‘Nansen’ lines contained at least one mutant allele. For ‘Desirée’ the percentage of lines with mutations in at least one allele were 90% (G2) and 74% (G4), while a mutation in all four alleles was observed in 55% and 47%, respectively, of the lines for both guide RNAs (Table 1). Examples from our HRFA chromatograms are available in Supplementary Material 6).
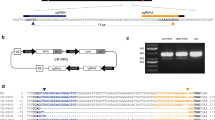
Lines producing yellow skinned tubers and with a putative full KO based on the HRFA screening, as well as two putatively non-edited lines (one for each of ‘Nansen’ and ‘Desirée’) producing red skinned tubers, were further sequenced for confirmation of induced mutations (Fig. 3).
Genotyping of full KO S. tuberosum lines with yellow potato tubers. Mutations as shown in individual alleles aligned to a corresponding wild-type (WT) fragment, determined by Sanger sequencing. Deleted nucleotides are shown with blue hyphens and inserted nucleotides are shown in bold red. (a) ‘Nansen’ line 177 17 mutated using G2. (b) ‘Nansen’ line 178 49 mutated using G4. (c) ‘Desirée’ line 179 16 mutated using G2. (d) ‘Desirée’ line 180 19 mutated using G4
Greenhouse growth and investigation of phenotype
Due to the necessity to apply for field trials for gene edited plants in Norway, we expected this to be a lengthy process, and probably would have been asked to conduct a step-by-step approach with greenhouse trials prior to field trials any way. Hence, we opted for contained greenhouse trials. However, this had some implications with regards to temperature during summer trials (Fig. 2), whereby we chose to perform the third clonal tuber generation period from January to April.
From protoplast transfection to harvest-ready plants in the greenhouse, via callus, shoot regeneration and transplanting, the timeline was 12 months (Fig. 4).
Development of putatively edited potato plants, from protoplasts to harvest-ready plants in the greenhouse. (a) Freshly isolated protoplasts visualized under light microscope. (b) 2-month old calli from individual protoplasts of ‘Nansen’. (c) regenerated shoots from 4-month old calli of ‘Desirée’. (d) regenerated shoot from one individual callus of ‘Nansen’ (e) Shoots transferred from in vitro to soil in greenhouse, kept in a plastic tent for acclimatization (f) Fully grown plants as watering was stopped and readied for harvest
Out of the 81 initial events (lines) transferred to pots, 77 survived the first summer trials and produced tubers. The foliage observed throughout the growing season appeared uniform, and typical for each cultivar, with no visual differences between the lines within a cultivar. However, we observed both white and light pink (WT) flower colour in the different lines, where the light pink flowers were also observed in partial KO lines (e.g., line 180 057 ‘Desirée’ G4), while the full KO lines (e.g., 179 054 ‘Desirée’ G2) had completely white flowers (Fig. 5bro.
Edited plants were obtained using both guide RNAs. These edited lines gave either full or partial KOs in both cultivars and yellow skinned tubers from both guide RNAs in both cultivars. Table 2 also displays that a majority of the full KO gave yellow skinned tubers, while the majority of the putatively non-edited lines gave red skinned tubers. It was also observed that even a KO of just 3 out of the 4 alleles resulted in a red skinned phenotype (e.g., ‘Desirée’ line 180,057) (Fig. 5). We also observe one mixed and one yellow line from G2 edits in ‘Nansen’ and one mixed line from G4 edits in ‘Desirée’ from Table 2.
. Figure 6 illustrates typical red, mixed and yellow skinned lines of ‘Nansen’.
Edited lines of S. tuberosum with induced mutations in the target sequence G2 in ‘Nansen’. To the left line 177,071, producing red skinned tubers, middle is line 177,017 with a typical mixed pot with both red and yellow skinned potatoes from one pot. To the right line 177,019, where yellow skin is obtained from original red ‘Nansen’ after editing. All lines are 1st clonal tuber generation trials in 2021
The second greenhouse clonal tuber generation was performed to investigate phenotype stability for yellow skin colour after gene editing. The yellow phenotype was retained for all full KO lines, while a red phenotype was retained for the unedited lines. Several lines showed an altered phenotype for other traits, such as elongation of the tuber and/or formation of knots on the main tuber (Fig. 7). This was not observed in the tubers in the first-year trials.
A third greenhouse clonal tuber generation investigated the effect of growing tubers with unusual phenotypes (knots on tubers) under more optimal potato growth conditions (lower temperatures). The third clonal tuber generation was under winter/spring conditions, rather than full summer, in contrast to the previous two years. When cultivated under lower temperatures from January-April, a clear reversal to first-clonal tuber generation phenotype was observed (Fig. 7) in all the lines.
Edited lines S. tuberosum with induced mutations in the target sequence G2 in ‘Nansen’ (177) and ‘Desirée’ (179). Putatively non-edited individuals for comparison retained a red phenotype in all three clonal tuber generations, while the edited lines retained the altered, yellow phenotype in the tubers. Note unusual phenotype in 2022, especially for the edited ‘Desirée’ line 179 14. Lower yields in the third clonal tuber generation (2023) is due to a shorter growth period (11 weeks), compared to the first two clonal tuber generations (23 weeks and 19 weeks). The numbering lacks one zero, e.g., clone 177 71 = 177,017
Discussion
This paper shows a proof-of-concept for CRIPSR/Cas9 editing of potato using two different gRNAs independently preassembled with Cas9 and transfected to isolated protoplasts. We have a long-term goal to use these positive results to edit other vital traits, such as disease resistance, in potato. Disease resistance has previously been demonstrated using conventional GMO technologies with three stacked genes for late blight in potato (Jones et al. 2014). We show here that the two gRNAs were independently successfully applied to two red skinned potato varieties, producing several lines with the expected yellow skinned tubers. We edited two different potato varieties, ‘Desirée’ which is a common variety to use in cell- and tissue culture work because it is responsive to regeneration from protoplast (González et al. 2020). ‘Nansen’ which is a new (2018) Norwegian variety, which is relatively well protected from late blight infection (https://graminor.no/sort/nansen/). However, ‘Nansen’ has not been subjected to protoplast regeneration previously. Both cultivars responded well to protoplast regeneration combined with gene editing, resulting in yellow skinned tubers.both cultivars. The breeding company, Graminor, has thus obtained a more desired skin colour in one of their own potato varieties.
The phenotypic screening revealed a colour change from red to yellow in the majority of the full KO plants, with yellow colouration in either all tubers or a mix of red and yellow tubers (Fig. 7). The exception was for G4 in ‘Desirée’, where tubers from three lines with in-frame mutations still appeared red, like the wild type tubers. It can be speculated that the in-frame mutations at G4 in ‘Desirée’ have no or only minor effect on the resulting protein structure and functionality. For the lines with partial KO (1–3 alleles), the tubers were either red, or a mix of red and yellow tubers. There was no visual difference between red tubers with partial KO and red tubers from control plants without editing observed in any allele. It is evident from Table 2 that to achieve a yellow skinned line, there must be a KO of all four alleles. This indicates that the presence of a single functional allele is enough for sufficient anthocyanin production with F3H, without clear additive effects of having several intact alleles. These findings are comparable to RNAi silencing of HMA4 homeologs in tobacco, where one functional HMA4 allele was sufficient to maintain wild-type levels of cadmium in the leaves (Liedschulte et al. 2017). For putatively non-edited individual plants for comparison, one line appeared yellow, while two lines had a mixed phenotype. This could be due to the selection of putatively non-edited plants based on the preliminary HRFA screening, where e.g., base substitutions leading to premature stop codons will appear similar to unedited lines, but which could have been edited all the same.
In this study, we hypothesized that CRISPR gene editing could be used for disruption of F3H in the anthocyanin biosynthesis (Fig. 1), which proved to be an efficient method for altering skin colour in red skinned potato tubers. Two primary phenotypes were observed, namely wild-type red or edited yellow. Several of the edited lines (20%) produced tubers with a mix of these phenotypes from a single protoplast-derived plant (Table 1). This also occurred in two of the lines in the “control” group. ‘Nansen’ was more prone to this mixture of genotypes, and this was found in 11 lines, compared to ‘Desirée’ where it was found in only two lines. There are at least two possible explanations for this mixture of skin colour derived from protoplast regeneration in our experiments. Despite protoplasts being the preferred method to ensure that plants originate in most cases from a single cell, in some cases (Reed and Bargmann 2021) the plants may have originated from more than one cell, resulting in putative chimeras. These chimeras could separate in the next clonal tuber generation and may be the reason for such mixture of skin colours from one single potato plant (Broertjes and Van Harten 1988). Another explanation could be somaclonal variation. We base this hypothesis on the fact that ‘Nansen’ reveals mixtures of tuber phenotypes independent of gRNA used, even though the lines are out-of-frame full KO, based on preliminary HRFA screening. The obtained mixtures of phenotype from these plants may be explained by changes occurring in the gene after the HRFA screening. These plants were cultivated in vitro and, especially for potato, we can expect somaclonal variants (Adly et al. 2023). It is noteworthy that no intermediate skin colour (e.g. lighter red) was observed in the lines with a partial knock-out. This indicates that F3H acts as a dominant gene with regards to skin colour in potato tubers.
The second and third clonal tuber generation trials gave us very valuable results. We wanted to ensure that the edited yellow skin colour was stable in the subsequent clonal tuber generations. Indeed, we found that the yellow tuber phenotype observed in the first clonal tuber generation greenhouse trial was retained for the second and third clonal tuber generation. This indicates stability of the altered phenotype, making gene editing a viable additional tool in the toolbox for future precision plant breeding in ‘Nansen’ when the goal is to change one or a few traits, while keeping most, or all, of the cultivar’s other traits intact. We have not recorded yields in these experiments in buckets in the greenhouse. The yield needs to be performed in a field trial, which we have not applied for, due to time constraints. Buckets yield is not representative compared to field trials and greenhouse conditions can be very different from field conditions with regards to both temperature and humidity.
When evaluating the results from the second clonal tuber generation of edited tubers, several lines showed signs of abnormal growth, such as tuber elongation and knots on the tubers. This was particularly prevalent in ‘Desirée’. One possibility is that this was the result of protoplast regeneration, which may induce widespread genome instability (Fossi et al. 2019). However, the mutated plants appeared to be of the expected phenotype in the first trials. Another possible explanation for the altered tuber phenotype could be due to the growth conditions in the greenhouse, with temperatures reaching 40 °C (Fig. 2). High temperatures have previously been seen to inhibit starch formation in potato tubers (Mohabir and John 1988), and any temperature stress could lead to excessive sugar accumulation (Kumar et al. 2004). The third clonal tuber generation in the greenhouse was therefore performed at a time of year (January-April) when it is easier to maintain optimal temperatures for potato inside a greenhouse (contained use according to Norwegian law). When given a more optimal growth temperature, the third clonal tuber generation reverted to the normal phenotype. This was true also when we selected to cultivate the strangest looking tubers as the seed potatoes. This indicates that greenhouse condition was a plausible cause for the second clonal tuber generation phenotypes (Fig. 7) and not an off-target effect showing up in the second trial. This demonstrates, however, the importance of successive generation trials after gene editing, just as one would do in a traditional plant breeding programme for potato.
The most common way to induce targeted gene mutations in potato is through protoplast editing. However, protoplasts are vulnerable cells without a cell wall, and have a complicated dedifferentiation and differentiation cycle before regeneration into novel shoots. There they may be more prone to spontaneous somaclonal variations than other types of plant material. According to Eeckhaut et al., protoplasts may be the ideal targets for CRISPR/Cas-mediated genome editing (Eeckhaut et al. 2020). They claim that this is particularly true when combined with DNA-free approaches including RNP, as was also mentioned earlier by (Woo et al. 2015). These may, for instance, result in variations among and between regenerants from a given genotype and a single regeneration experiment that are not issued from genome editing, but rather from somaclonal variation. Potato is known to be more vulnerable than most crops through all types of adventitious shoot regeneration, and cell- and tissue culture is frequently used as a method of creating variation in a breeding program (Larkin and Scowcroft 1981). Adly et al. (2023) recently reported use of somaclonal variation to produce potato clones with an altered starch accumulation. Therefore, we were cautious of the abnormal looking potatoes obtained in the second clonal tuber generation. However, as mentioned, the third-clonal tuber generation trials under a more favourable time of year for potato in greenhouses, revealed that the most likely cause of the frequent knots on (particularly) the ‘Desirée’ potato, was due to elevated temperatures in critical phases of the tuber development in our experiments. Still, from Table 2, we can observe a certain degree of variation in the putatively non-edited plants, for G2 in ‘Nansen’ and G4 in ‘Desirée’. We can not exclude that this is due to somaclonal variation in our experiments.
The successful edit of skin colour in ‘Nansen’ and ‘Desirée’ provided a change in an easily observable phenotype, while also providing protocols suitable for ‘Nansen’. This could form the basis for continued research to improve more complex traits such as disease resistance. Of these, the most obvious candidate for editing is late blight resistance, one of the most devastating diseases in potato and the cause of the Irish potato famine in the 1840s (Mizubuti and Fry 2006). One possible approach is to adapt KO of putative susceptibility genes, which has shown promise in both Arabidopsis (Karmakar et al. 2022) and potato (Kieu et al. 2021; Sun et al. 2016) when using either CRISPR/Cas9 or RNAi technology.
A common regulatory obstacle of gene technology is the introduction of foreign DNA into the genome. This is prevented by combining protoplasts with ribonucleoprotein complexes, which has been shown to efficiently produce transgene-free edits in various species through DNA-free gene editing (Andersson et al. 2018; Cho et al. 2013; Kim et al. 2014; Liang et al. 2017; Woo et al. 2015). If common transformation approaches are used, such as Agrobacterium-mediated transformation of vectors for expression of the CRISPR tools, it will usually incorporate foreign DNA into the plant genome. The edited plants will then be considered traditional GMOs in the regulatory sense, unless foreign DNA can be removed by backcrossing or other approaches (Parisi and Rodríguez-Cerezo 2021). To attempt this in potato, would be contradictory to the purpose of using CRISPR to alter single traits in potato, as severe inbreeding depression makes backcrossing challenging. Also, the crossing of genotypes would result in a recombination of the genes, thereby altering the next clonal tuber generation also in many of their desirable traits. The advantage of using modern technology would then be reduced to simply introducing the novel traits into the gene pool, in the same way as it is used in seed propagated crops. The advantage of vegetatively propagated crops; once the desired phenotype/genotype is selected, is that the genotype can be preserved as a clonal cultivar. Editing an existing cultivar, or a selected clone from a breeding program, and altering the one (or few) traits which need improvement, is therefore an appealing idea.
Furthermore, transgene-free GE plants are either unregulated or less strictly regulated than traditional GMOs in many parts of the world, such as the United States of America, Canada, Australia, Japan and several South American countries (Schmidt et al. 2020). Additionally, there is an ongoing discussion both in Norway and the EU on regulation of crops developed through gene technology. One of the proposals was to have a tier-based regulation, where transgenic plants are regulated as in the current system, while transgene-free, gene-edited crops are regulated less strictly and closer to traditional plant breeding (Bratlie et al. 2019). The Norwegian government has recently (June 6th 2023) received an Official Norwegian Report (NOU 2023:18, 2023) where the Committee majority suggested that changes in the regulatory process could deal with precision breeding and cis-genesis in the future. Similarly, EU has proposed (Comission, 2023) that plants developed using new genomic techniques, where plants that cannot be easily distinguished from those of conventionally bred plant varieties, could be regulated as conventionally bred varieties. On the other hand, the Commission proposes that plants with more than 20 modified nucleotides should be regulated like “conventional” GM plants. This proposal currently has been subjected to a public hearing, and the last word has not been said. Being able to produce GE plants that are unregulated or less strictly regulated than current GMOs can be crucial for local adaptation, as the cost of approving GM food or feed in the EU has been estimated to somewhere between €11 and €16.7 million (EuropaBio 2019). As such, a transgene-free approach might be the only way for publicly funded universities or small and medium sized enterprises to bring valuable gene edited crops to the market. Our findings with potato demonstrate successful adaptation of existing protocols of transgene free CRISPR/Cas editing to a commercially interesting Norwegian cultivar.
Conclusions
By growing the edited potato lines over several regenerated cycles, we demonstrated that unexpected phenotypes might arise in one clonal tuber generation, while the previous and subsequent clonal tuber generations appear as expected. This is an important result to bear in mind for two reasons: (1) when phenotyping putative gene edited plants, every care should be taken to ensure optimal growing conditions for the species in question, and (2) when testing gene edited plants, more than one clonal tuber generation should be grown, to determine the stability of the editions.
This demonstrates that multiple events, multiple climatic conditions and careful selection for the superior genotype is equally valid subsequent to using gene edited methods, as it has been for the past century for traditional plant breeding.
This publication forms the basis for tools for precision breeding in Norwegian potato cultivars with the aim of providing Norwegian farmers with a notably increased resistance towards our largest disease for potatoes in the world; late blight (Phytophthora infestans), costing the Norwegian farmers more than 122 million NOK annually (Forbes et al. 2023). With this goal in mind, forthcoming studies will focus on the identification of the putatively best genes to be edited to achieve a sustainable production with a minimum use of chemicals.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adly WMRM, Niedbała G, EL-Denary ME, Mohamed MA, Piekutowska M, Wojciechowski T, Abd El-Salam E-ST, Fouad AS (2023) Somaclonal Variation for Genetic Improvement of Starch Accumulation in Potato (Solanum tuberosum) Tubers. Plants 12 (2): 232
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Andersson M, Turesson H, Nicolia A et al (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36:117–128. https://doi.org/10.1007/s00299-016-2062-3. )
Andersson M, Turesson H, Olsson N, Fält AS, Ohlsson P, Gonzalez MN, Samuelsson M, Hofvander P (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164(4):378–384
Bethke PC, Nassar AM, Kubow S, Leclerc YN, Li X-Q, Haroon M, Molen T, Bamberg J, Martin M, Donnelly DJ (2014) History and origin of Russet Burbank (netted gem) a sport of Burbank. Am JPotato Res 91:594–609
Bonierbale MW, Amoros WR, Salas E, Jong W (2020) Potato breeding. The potato crop. Springer, Cham, pp 163–217
Bratlie S, Halvorsen K, Myskja BK, Mellegård H, Bjorvatn C, Frost P, Heiene G, Hofmann B, Holst-Jensen A, Holst‐Larsen T (2019) A novel governance framework for GMO: a tiered, more flexible regulation for GMO s would help to stimulate innovation and public debate. EMBO Rep 20(5):e47812
Broertjes C, Van Harten AM (1988) Applied mutation breeding for vegetatively propagated crops. 1st Edition Elsevier, Amsterdam
Cho SW, Lee J, Carroll D, Kim J-S, Lee J (2013) Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics 195(3):1177–1180
Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean, Irina M, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R et al (2021) Ensembl 2022. Nucleic Acids Res 50(D1):D988–D995. https://doi.org/10.1093/nar/gkab1049
Devaux A, Kromann P, Ortiz O (2014) Potatoes for sustainable global food security. Potato Res 57(3):185–199
EuropaBio (2019) Pricing innovation out of the EUPricing innovation out of the EU (accessed: 25/05/2023)
European Commission, REGULATION OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL on (2023) Proposal for a plants obtained by certain new genomic techniques and their food and feed, and amending Regulation (EU) 2017/625 COM (2023) 411 final
FAOSTAT (2023) https://www.fao.org/faostat/en/#data/FBS (accessed: 05.07.2023)
Flaten O, Hisano S (2007) Food security policy in a food importing country: the case of Norway. Nougyou Keizai (Agriculture Economy) 73(8):129–136
Forbes E, Wulff-Vester AK, Hvoslef-Eide TAK (2023) Will genetically modified late blight resistant potatoes be the first GM crops to be approved for commercial growing in Norway? Front Plant Sci 14:1137598. https://doi.org/10.3389/fpls.2023.1137598
Fossi M, Amundson K, Kuppu SBritt A, Comai L (2019) Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol 180(1):78–86
Gemenet DC, Khan A (2017) Opportunities and challenges to implementing genomic selection in clonally propagated crops. Genomic Selection for Crop Improvement, pp 185–198
Ghimire M (2022) 10 + Popular Types of Potatoes in UK. https://plantscraze.com/types-of-potatoes-uk/#:~:text=Maris%20Peer%20is%20one%20of%20the%20most%20famous,types%20after%20Maris%20Piper%20in%20the%20UK.%202. (accessed: 02.05.2023)
González MN, Massa GA, Andersson M, Turesson H, Olsson N, Fält A-S, Storani L, Décima Oneto CA, Hofvander P, Feingold SE (2020) Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front Plant Sci 10:1649
Holton TA, Tanaka Y (1994) Blue roses: a pigment of our imagination? Trends in biotechnology (regular. 12(2):40–42
Jansky SH, Spooner DM (2018) The evolution of potato breeding. Plant Breed Reviews 41:169–214
Jones JDG, Witek K, Verweij W, Jupe F, Coke D, Dorling S, Tomlinson L, Smoker M, Perkins S, Foster S (2014) Elevating crop disease resistance with cloned genes. Phil Trans R Soc B369:20130087. https://doi.org/10.1098/rstb.2013.0087
Karmakar S, Das P, Panda D, Xie K, Baig MJ, Molla KA (2022) A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Sci 323:111376
Kieu NP, Lenman M, Wang ES, Petersen BL, Andreasson E (2021) Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci Rep 11(1):1–12
Kim S, Kim D, Cho SW, Kim J, Kim J-S (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24(6):1012–1019
Kumar D, Singh B, Kumar P (2004) An overview of the factors affecting sugar content of potatoes. Ann Appl Biol 145(3):247–256
Larkin PJ, Scowcroft WR (1981) Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:197–214
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8(1):1–5
Liedschulte V, Laparra H, Battey JND, Schwaar JD, Broye H, Mark R, Klein M, Goepfert S, Bovet L (2017) Impairing both HMA4 homeologs is required for cadmium reduction in tobacco. Plant Cell Environ 40(3):364–377
Mishra T, Luthra SK, Raigond P, Kaundal B (2020) Anthocyanins: Coloured Bioactive Compounds in Potatoes. In Raigond P, Singh B, Dutt S, Chakrabarti SK (eds) Potato: Nutrition and Food Security:173–189. Springer Singapore
Misra AK (2014) Climate change and challenges of water and food security. Int J Sustainable Built Environ 3(1):153–165
Mizubuti ES, Fry WE (2006) Potato late blight. The epidemiology of Plant diseases. Springer, pp 445–471
Mohabir G, John P (1988) Effect of temperature on starch synthesis in potato tuber tissue and in amyloplasts. Plant Physiol 88(4):1222–1228
Nicolia A, Falt A-S, Hofvander P, Andersson M (2021) Protoplast-based method for genome editing in tetraploid potato. Methods Mol Biol 2264:177–186 Clifton, N.J.)
NOU (2023) Genteknologi i en bærekraftig fremtid. Norges Offentlige Utredninger (NOU) 2023:18
Parisi C, Rodríguez-Cerezo E (2021) Current and future market applications of new genomic techniques. Publications Office of the European Union, Luxembourg: JRC123830
Potet F (2021) Nansen. https://potet.no/produkter/nansen (accessed: 10.05.2023)
Reed KM, Bargmann BOR (2021) Protoplast Regeneration and its use in new plant breeding technologies. Front Genome Ed 3:734951. https://doi.org/10.3389/fgeed.2021.734951
Schmidt SM, Belisle M, Frommer WB (2020) The evolving landscape around genome editing in agriculture: many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep 21(6):e50680
Singh BK, Delgado-Baquerizo M, Egidi E, Guirado E, Leach JE, Liu H, Trivedi P (2023) Climate change impacts on plant pathogens, food security and paths forward. Nat Rev Microbiol. https://doi.org/10.1038/s41579-023-00900-7
Slater AT, Cogan NO, Hayes BJ, Schultz L, Dale MFB, Bryan GJ, Forster JW (2014) Improving breeding efficiency in potato using molecular and quantitative genetics. Theor Appl Genet 127(11):2279–2292
Sun K, Wolters A-MA, Vossen JH, Rouwet ME, Loonen AE, Jacobsen E, Visser RG, Bai Y (2016) Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res 25(5):731–742
Wheeler T, Von Braun J (2013) Climate change impacts on global food security. Science 341(6145):508–513
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim S-T, Choe S, Kim J-S (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33(11):1162–1164
Wu M, Liu H, Lin Y, Chen J, Fu Y, Luo J, Zhang Z, Liang K, Chen S, Wang F (2020) In-frame and frame-shift editing of the Ehd1 gene to develop japonica rice with prolonged basic vegetative growth periods. Front Plant Sci 11:307
Zhao X, Jayarathna S, Turesson H, Fält A-S, Nestor G, González MN, Olsson N, Beganovic M, Hofvander P, Andersson R (2021) Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci Rep 11(1):4311
Acknowledgements
The authors thank Dag-Ragnar Blystad and Sissel Haugslien from the Norwegian Institute of Bioeconomy (NIBIO), Ås, Norway for providing disease-free plants of ‘Nansen’. We are grateful to Mark Smedley of John Innes Centre, Norwich, UK for his guidance and tutoring on GuideRNA assembly. We thank Tone Melby and Pelle Mikkelsen for their valuable technical assistance in the Plant Cell Laboratory (NMBU), and to Gry Skjeseth and Silje Roksti for the maintenance of the potato plants in the greenhouse of the Norwegian University of Life Sciences (NMBU), Ås, Norway. We are also acknowledging technical support from Ann-Sofie Fält, Niklas Olsson and Helle Turesson at the Swedish University Agricultural Sciences (SLU), Alnarp, Sweden and the technical staff from Graminor Ltd, especially Anja Haneberg and Bjarne Kjøs. The work was performed as part of the GENEinnovate (Project # 281928) co-funded by the Norwegian Research Council (RCN) and Graminor Breeding Company, Hamar, Norway. The Norwegian University of Life Sciences (NMBU) provided a full PhD stipend to Anders Wulff-Vester.
Funding
The study was a part of the project “GENEinnovate” funded by the Norwegian Research Council (Project # 281928), plant-, aqua and animal breeding companies in Norway (Norsvin, GENO; Aquagen and Graminor) and a PhD stipend to Anders-Wulff-Vester from the Norwegian University of Life Sciences (NMBU) in Ås, Norway.
Open access funding provided by Norwegian University of Life Sciences
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Anders Wulff-Vester, Mariette Andersson, Per Hofvander, Trine Hvoslef-Eide, and Muath Alsheikh. The first draft of the manuscript was written by Anders Wulff-Vester. Mariette Andersson provided sections of the Mat Meth and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Joyce Van Eck.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wulff-Vester, A., Andersson, M., Brurberg, M.B. et al. Colour change in potato (Solanum tuberosum L.) tubers by disruption of the anthocyanin pathway via ribonucleoprotein complex delivery of the CRISPR/Cas9 system. Plant Cell Tiss Organ Cult 157, 25 (2024). https://doi.org/10.1007/s11240-024-02743-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11240-024-02743-3