Abstract
The CRISPR/Cas9 system is a remarkably promising tool for targeted gene mutagenesis, and becoming ever more popular for modification of ornamental plants. In this study we performed the knockout of flavonoid 3′-hydroxylase (F3′H) with application of CRISPR/Cas9 in the red flowering poinsettia (Euphorbia pulcherrima) cultivar ‘Christmas Eve’, in order to obtain plants with orange bract colour, which accumulate prevalently pelargonidin. F3′H is an enzyme that is necessary for formation of cyanidin type anthocyanins, which are responsible for the red colour of poinsettia bracts. Even though F3′H was not completely inactivated, the bract colour of transgenic plants changed from vivid red (RHS 45B) to vivid reddish orange (RHS 33A), and cyanidin levels decreased significantly compared with the wild type. In the genetically modified plants, an increased ratio of pelargonidin to cyanidin was observed. By cloning and expression of mutated proteins, the lack of F3′H activity was confirmed. This confirms that a loss of function mutation in the poinsettia F3′H gene is sufficient for obtaining poinsettia with orange bract colour. This is the first report of successful use of CRISPR/Cas9 for genome editing in poinsettia.
Key message
Genome edited poinsettia plants show loss-of-function mutations in the F3’H gene introduced by CRISPR/Cas9. This is sufficient to change the bract colour from red to reddish orange.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The winter-flowering Euphorbia pulcherrima (poinsettia or Christmas Star) belongs to the most economically important potted ornamental plants, especially during the Christmas season (Ecke 2004). Traditionally, the mass market prefers intense scarlet or dark-red colouration of the bracts (Taylor et al. 2011), the latter being leaves that change their colour from green to red to support the plain cymes in pollinator attraction. However, during the last two decades, a huge diversity of red hues and novelty colours and styles has arisen in the poinsettia assortment, because a considerable number of consumers is willing to pay higher prices for unusual varieties (Barrett 2005). Among these, orange-red bract colour seems to attract European and North-American consumers, and has the potential to extend the market e.g. as a Halloween speciality (Barrett 2005).
The pigments responsible for poinsettia bract colouration, at least for the red hues, are the anthocyanins, a well-known group of secondary metabolites (Stewart et al. 1979; Slatnar et al. 2013). Two main types of anthocyanins can be distinguished in poinsettia, based on the number of hydroxyl groups in the B-ring, the pelargonidin type (one hydroxyl group) and the cyanidin type (two hydroxyl groups). Recently, we have shown that the rare orange-red bract colouration of poinsettia (Euphorbia pulcherrima) is associated with a prevalence of pelargonidin-type anthocyanins and a somewhat decreased anthocyanin concentration in general. We also reported that accumulation of pelargonidin-based pigments in poinsettia is caused by different mechanisms, of which one is a strong reduction of flavonoid 3′-hydroxylase (F3′H) activity in the bracts (Nitarska et al. 2018). F3′H (EC 1.14.13.21) is a membrane bound enzyme associated with the cytosolic site of the endoplasmic reticulum (Schuler and Werck-Reichhart 2003) and belongs to the subfamily CYP75B of cytochrome P450-dependent monooxygenase (P450) (Tanaka 2006; Chapple 1998). F3′H is responsible for the introduction of a second hydroxyl group in the B-ring thereby leading to the formation of cyanidin type anthocyanins (Fig. 1a). F3′H, however, does not act on anthocyanins themselves, but on one of the intermediates upstream of the anthocyanin forming step (Schwinn et al. 2014). Particularly, the number of hydroxyl groups in the B-ring of the dihydroflavonol precursors determines the colour of each group of anthocyanins. Dihydrokaempferol (DHK), with one hydroxyl group, is converted to orange-red pelargonidin type anthocyanins, and dihydroquercetin (DHQ), with two hydroxyl groups, to red-pink cyanidin type (Halbwirth 2010) (Fig. 1). As has been described (Nakatsuka et al. 2007), silencing of F3’H is an important factor for obtaining plants that accumulate prevalently pelargonidin type anthocyanins in tobacco.
Simplified overview of the anthocyanin pathway. Abbrev: ANS: anthocyanidin synthase, CHI: chalcone isomerase, DFR: dihydroflavonol 4-reductase, FHT: flavanone 3-hydroxylase, F3′H: flavonoid 3′-hydroxylase, F3′5′H: flavonoid 3′,5′-hydroxylase. a: The prevalent pathway in naturally occurring poinsettia; b: The chosen strategy for attaining reddish orange bracts
Recently, the Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR/Cas9) system was reported as a promising tool for targeted genome modification in plants (Corte et al. 2019) and it has quickly developed into the most widely used genome editing method (Kishi-Kaboshi et al. 2018). In this system, double-strand breaks (DSBs) are introduced by Cas9 nuclease, guided by a 20 nucleotide sequence, single guide RNA (sgRNA) (Jinek et al. 2012). There are two mechanisms of DSB repair, homology recombination (HDR) and non -homologous end joining (NHEJ) (Puchta 2005). The second mechanism usually leads to the introduction of a mutation, insertion or deletion, in the gene sequence and consequently to a loss of function in the protein (Chiruvella et al. 2013). This approach is now commonly used for specific gene knockout in many plant species (Ma et al. 2016). For the first time, the CRISPR/Cas9 system was used in ornamentals for silencing phytoene desaturase (PDS) in petunia (Petunia x hybrida) (Zhang et al. 2016). The first successful alteration of flower traits was reported in Japanese morning glory (Ipomoea nil), where dihydroflavonol 4–reductase (DFR) was targeted (Watanabe et al. 2017). Currently this approach is becoming more and more popular for the modification of ornamentals. Up to now it was used for targeting multiple secondary metabolism genes in orchid (Phalaenopsis) (Tong et al. 2020), chrysanthemum (Chrysanthemum morifolium) (Kishi-Kaboshi et al. 2017), lily (Lilium pumilum, Lilium longiflorum) (Yan et al. 2019), torenia (Torenia fournieri) (Nishihara et al. 2018) and gentian (Gentiana triflora × Gentiana scabra) (Tasaki et al. 2019).
This study is a proof of concept for the conclusion from our previous work (Nitarska et al. 2018), that a mutation in the F3′H gene is sufficient to obtain pelargonidin accumulating poinsettia. We attempted to knockout the poinsettia F3′H gene by applying the CRISPR/Cas9 method to obtain poinsettias prevalently accumulating pelargonidin type anthocyanins in their bracts (Fig. 1b). To our knowledge, this is the first report of a genome-edited poinsettia.
Materials and methods
Chemicals
(2-14C)-Malonyl-coenzyme A (55 mCi mmol−1) was purchased from New England Nuclear Corp. GmbH (Vienna, Austria). Synthesis of radiolabeled substrates was performed as described in (Halbwirth et al. 2006). Reference substances for HPLC analysis (cyanidin, pelargonidin, delphinidin, malvidin, petunidin,) were purchased from Extrasynthese (Genay, France).
Plant material
Poinsettia Euphorbia pulcherrima cultivar ‘Christmas Eve’ (Klemm + Sohn GmbH & Co. KG, Germany) was used for transformation. Plants were grown in the greenhouse under long day conditions (16 h day/8 h night). For Agrobacterium-mediated transformation internode stem explants were used. Excised stems were surface sterilized by washing for 10 min in 1.5% solutions of NaOCl with 1 drop of Tween 20 and then washed twice for 10 min in sterile water. In the next step, internode stems were cut (around 1 mm thick) and placed on callus induction media [CIM–MS media supplemented with 0.2 mg L−1 4-chlorophenoxy acetic acid (CPA) and 0.2 mg L−1 6-Benzylaminopurine (BAP)] for 4–5 days and then explant discs were used for transformation.
sgRNA design and cloning
For transformation, the binary vector pDe-Sa_Cas9 (Steinert et al. 2015), carrying a kanamycin resistance gene (nptII) to facilitate selection was used (Fig. 2). A sgRNA sequence was designed based on the sequences available in the Gene Bank (KY273440.1), with application of the online tool CHOPCHOP (Labun et al. 2016; Montague et al. 2014). A 20 nucleotide long sgRNA sequence (CAGTCAATAGCCTCCTTGGC), without PAM sequence (TCGGGT), was cloned into a pEN-Sa_Chimera vector (primers for cloning: pEN-Sa_Chim_EpF3HgRNA_F ATTGCAGTCAATAGCCTCCTTGGC and pEN-Sa_Chim_EpF3HgRNA_R AAACGCCAAGGAGGCTATTGACTG) (Steinert et al. 2015). In the second step, a sgRNA expression cassette was transferred to the destination vector (pDe-Sa_Cas9) by single-site Gateway LR reaction (Thermo Fisher Scientific https://www.thermofisher.com/pl/en/home/life-science/cloning/gateway-cloning/protocols.html). Cloning success was confirmed by sequencing by a commercial supplier (LGC, Germany).
Transformation and regeneration
Agrobacterium tumefaciens strain GV3101, carrying the pDe-Sa_Cas9 vector with cloned sgRNA was cultivated in SOB media (2% w/v tryptone, 0.5% w/v yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4) supplemented with 50 mg L−1 rifampicin, 100 mg L−1 spectinomycin and 30 mg L−1 gentamicin for 24 h with shaking (200 rpm) at 28 °C. Then 5 mL of this culture was used to inoculate 50 ml of Minimal A medium (10.5 g L−1 K2HPO4, 4.5 g L−1 KH2PO4, 1 g L−1 (NH4)2SO4, 0.52 g L−1 Na3C6H5O7 × 2 H2O, 0% w v−1 glucose, 0.005% MgSO4 × 7 H20, 0.00025% thiamine) and was further cultivated at identical conditions till OD reached 0.5. Then bacteria were used for transformation. Explants were incubated with 10 mL of Agrobacterium inoculum for 30 min with gentle shaking and dried on sterile paper, then placed on the CIM for 2 days of co-cultivation. As control explants were incubated with 10 mL of Minimal A medium. Subsequently, discs were washed in sterile water containing the antibiotics 250 mg L−1 cefotaxim and 150 mg L−1 timentin for 30 min and dried on sterile paper, then placed on the CIM supplemented with 250 mg L−1 cefotaxim and 150 mg L−1 timentin for callus induction. After 14 days on CIM, explants were transferred to somatic embryo induction media [SEIM–MS medium with 0.2 mg L−1 1-Naphthaleneacetic acid (NAA) and 0.1 mg L−1 isopentenyl adenine (2ip)] supplemented with 250 mg L−1 cefotaxim, 150 mg L−1 timentin and 2.5 mg L−1 kanamycin, for induction of somatic embryogenesis. After 3–6 weeks, when somatic embryos were visible, explants were transferred to somatic embryo maturation medium (SEMM–MS medium with 0.05 mg L−1 BAP) supplemented with 250 mg L−1 cefotaxim, 150 mg L−1 timentin and 200 or 50 mg L−1 kanamycin for further cultivation and selection. Fully regenerated plants were placed on MS media without antibiotics, propagated and transferred to the greenhouse. Plants were transferred from the medium into paper-wrapped substrate plugs and carefully adapted to greenhouse conditions. After hardening, plugs were potted in 1 L-pots containing Einheitserde P substrate (Hermann Meyer KG, Germany) and grown with an average temperature of 22 °C under natural light conditions. Three cuttings per line were grafted onto Phytoplasma-containing rootstock to promote branching of the scion. One successfully grafted scion per line was further developed as mother plant to generate cuttings for vegetative propagation. At least 5 cuttings per line were rooted and potted into 1 L-pots containing Einheitserde P substrate (Hermann Meyer KG, Germany). They were cultivated with an average temperature of 22 °C under short-day conditions (9 h of daylight, 15 h of darkness) to induce flower formation and to stimulate the development of colored bracts.
Screening of regenerated plants
Genomic DNA was extracted from leaves of regenerated poinsettia with InVisorb Plant mini kit (Stratec, Germany). Transgenic plants were selected by PCR, where presence of nptII, cas9 and sgRNA sequences was detected (primers’ sequences are provided in Suppl. Table S1). For screening, GoTaq polymerase (Promega, Germany) was used. The reaction contained a final volume of 20 µL: 4 µL 5 × GoTaq Green buffer, 0.4 µL dNTPs, 1 µL forward primer (10 µM), 1 µL reverse primer (10 µM), 2 µL DNA, 0.2 µL Taq; reaction conditions: 2 min 94 °C initial denaturation, 40 cycles (98 °C, 30 s.denaturation, 60/62 °C, 30 s. primer annealing, 72 °C, 45 s. extension), 10 min 72 °C final extension. From positive plants, the total RNA was extracted by mirPremier Kit (Sigma Aldrich, Austria) according to the protocol provided. cDNA was synthesized with RevertAid H Minus Reverse Transcriptase (Thermo Scientific, US) according to the manufacturer’s protocol. Primers and PCR conditions were the same as described above. Integrity of DNA as cDNA was confirmed by amplification of poinsettia endogenous genes (actin, GAPDH or EpF3′H fragment).
Sequence analysis
Genomic DNA from genome-edited plants’ leaves was extracted using DNeasy plant mini kit (Qiagen, Germany) according to the manufacturer’s protocol. Gene fragments were amplified with EpF3′HpYes-F and EpF3′H-crispr-R primers (Suppl. Table S1) with Q5 High Fidelity DNA Polymerase (New England Biolabs, Austria). PCR program: 30 s. 98 °C for initial denaturation, 30 cycles (98 °C 30 s. denaturation, 62 °C 30 s. primer annealing, 72 °C 30 s. extension), 2 min 72 °C final extension. PCR products were separated on 1% agarose and extracted using Wizard SV Gel and PCR Clean-up System (Promega, Germany). Sub-libraries were constructed with application of NEBNext Ultra II DNA Library Prep kit for Illumina (New England Biolabs, Austria) according to the manufacturer’s protocol. For indexing, NEBNext Multiplex Oligos for Illumina (Index Primer Set 1) (New England Biolabs, Austria) were used. Concentration of each sub-library was measured with Qubit (Invitrogen, USA). The bulked library was obtained by mixing equal amounts of each sub-library. The concentration of the bulked library was measured with Qubit and checked on Fragment Analyzer (Agilent, USA), and then it was diluted to 8 pM. As a control PhiX (Illumina, USA) was spiked into bulked library in a concentration of 1.3%. Sequencing runs were performed on Illumina MiSeq system, with application of MiSeq v2 Reagent Kit 300 cycles (Illumina, USA) (2 × 150 reads). Sequencing results were analysed with CRISPResso2 software (Clement et al. 2019).
HPLC analysis
Poinsettia bract pigment analysis was performed by HPLC as previously described (Haselmair-Gosch et al. 2018). For the extraction, 0.5 g of shock frozen poinsettia bracts were used in 1.5 mL of 2 M HCl in methanol.
Gene expression studies
The F3′H and Cas9 gene expression was evaluated by qPCR using the StepOnePlus system (Applied Biosystems, Germany) and the Luna® Universal qPCR Master Mix (New England Biolabs, Austria) according to the manufacturers’ protocols. The analysis was performed in three independent replicates and the results were normalized to the two control genes, actin and translation elongation factor I alpha (EF1 A) (Zhang et al. 2013). Primer efficiency (Suppl. Table S1) and the relative expression ratio were calculated according to Pfaffl (Pfaffl 2004). Product specificity was confirmed by analysis of melting curves.
Poinsettia F3’H cloning and heterologous expression in yeast
F3′H gene from transgenic plants and WT were cloned into pYes2.1/V5-His-TOPO vector (Invitrogen, US) using EpF3′HpYes-F and EpF3′HpYes-R primers (Suppl. Table S1). Heterologous expression in yeast and the F3′H enzyme assay was performed as previously described (Nitarska et al. 2018).
Statistical analysis
The statistical analysis was performed in GraphPad Prims version 8.4.3 for Mac. Normality of data was checked with the Shapiro–Wilk test. Statistical significance was calculated using a double-tailed, unpaired t-test when the variances were equal or unpaired t-test with Welch’s correction when variances were different. The Mann–Whitney test was used for not normally distributed data.
Results
Transformation and regeneration
In order to obtain poinsettia plants with inactive F3′H, a CRISPR/Cas9 approach was used. Poinsettia stem segments were transformed via Agrobacterium-mediated transformation with the pDe-Sa_Cas9 vector carrying a sgRNA sequence specific to poinsettia F3′H (Fig. 2). For transformation, cultivar ‘Christmas Eve’ was selected as the best for regeneration among four other tested cultivars (data not shown). In total, seven experiments were performed, which resulted in the regeneration of 105 putatively transgenic plants (Table 1). Due to the fact that the stem segments originated from the greenhouse and the surface sterilization process was not always sufficient, a huge number of explants was lost during the first days after transformation, due to fungal and bacterial infections. Although the callus formation process was very efficient, the embryo formation could be observed only on a few explants. In experiments A to D, 200 mg L−1 of kanamycin was used for selection, which resulted in the regeneration of only 7 plants, because most of the embryos and plantlets did not survive the selection step. In order to obtain more regenerated plants in experiments E to G we lowered the kanamycin content to 50 mg L−1 as was suggested in literature (Clarke et al. 2008). At these conditions, we obtained an additional 98 putatively transformed plants, which would suggest that the previous selection conditions could have been too harsh for the cultivar ‘Christmas Eve’.
Screening of regenerated plants
Screening of regenerated plants was performed in two steps. In the first step, genomic DNA from leaves of regenerated plants was extracted and the presence of the transgene was investigated by PCR (targets: nptII, cas9). In step II, the total RNA was extracted from plants harbouring the transgene as identified in step I. Thereafter, cDNA was synthesized and used as a template for PCR (targets: nptII, cas9, gRNA) to check the expression of the transgene. In the first screening step, a total of 14 positive plants were found (Table 2, Fig. 3). The high number of negative plants (91) can be attributed to the decrease of the kanamycin concentration in the SEM media, which caused a bigger number of escapes. The second step of screening revealed that expression of the transgene could be detected only in three plants, which all originated from experiment B (200 mg L−1 kanamycin) (Fig. 4). Positive lines named B2, B158 and B284 were transferred to the greenhouse for further cultivation.
PCR evaluation of the transgenic plants and WT control on gDNA level. a: Amplification of EpF3’H fragment (primers EpF3’HpYesF and EpF3’HcrisprR), b: Amplification of cas9 fragment (primers Cas9-F1 and Cas9-R1). M molecular size standard 1 kb Plus DNA ladder (New England Biolabs, Austria), PC positive control (plasmid), NTC non template control. Selected fragments of ladder are labelled for better orientation
PCR evaluation of the transgenic plants and WT on cDNA level. a: Amplification of EpF3’H fragment (primers EpF3’HpYesF and EpF3’HcrisprR), b: Amplification of EpGAPDH fragment (primers EpGAPD-F and EpGAPDH-R), c: cas9 fragment (primers Cas9-F1 and Cas9-R1). M molecular size standard 1 kb Plus DNA ladder (New England Biolabs, Austria), PC positive control (plasmid), NTC non template control. Selected fragments of ladder are labelled for better orientation
Sequence analysis
To analyse the F3′H sequence of transgenic plants, next generation sequencing (NGS) was performed. 24% of the reads for line B2 reddish orange (B2.1) and 19% of line B2 chimera (B2.2) was modified (Table 3). For lines B158 and B284, almost no modifications were found. The most prevalent modification for line B2 was an insertion of a thymine (T) located three nucleotides before the PAM sequence. Nevertheless, in all transgenic lines, the most prevalent sequence was that of the WT, which suggests that the genome editing approach was not efficient enough. Even for line B2 more that 66% reads were identical to the sequence of the WT, which explains the presence of cyanidin type anthocyanins in the bracts. Interestingly, only a slightly higher number of WT reads was detected for line B2 chimeric plants in comparison to the fully reddish orange B2 plant. In all analysed plants, also other sequences than the major reads were present and typically consisted of 0.5% of total reads or less. This outcome is considered to be a result of PCR and NGS errors. Sequencing analysis confirmed that the genome editing in line B2 worked, but still the WT sequence is present, which results in just a partial shift toward pelargonidin in anthocyanin synthesis for this transgenic line.
Bract analysis
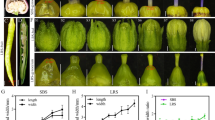
Bract colouration was induced by cultivation for 8 weeks at short day conditions (11 h day, 13 h night). In the case of line B2, two plants were obtained by propagation of the regenerated shoot, of which one showed bracts with brighter colour (hereafter named B2.1) compared with the wild type (WT) (Fig. 5a, b). The second plant (hereafter named B2.2) showed scattered points of brighter colour on the bracts, which could be described as chimeric phenotype (Fig. 5c). For lines B158 and B284, one plant each was obtained, which showed the same phenotype as the WT. All lines after first blooming were further propagated and analysed. The colour of propagated plants of line B2.1 was assigned to groups between 33A and 44B on the RHS charts (5th edition 2007), both are described as vivid reddish orange, but 33A is placed in the orange colour group and 44B in the red colour group. Wild type ‘Christmas Eve’ was assigned to group 45B, described as vivid red.
To analyse the anthocyanin content in the transgenic poinsettia bracts, HPLC analysis was performed. The total amount of anthocyanins in bracts was lower for line B2.1 with fully reddish orange bracts (Fig. 6a). The fully reddish orange plants of line B2.1 had a significantly lower amount of cyanidin compared with the WT (Fig. 6b). In all other lines, the cyanidin level was only slightly lower than in the WT (Fig. 6b). The pelargonidin amount, however, in almost all transgenic lines was not significantly different. Only for line B2.2 the increase of the pelargonidin content was observed (Fig. 6c).
Anthocyanins in bracts of poinsettia. a: Total anthocyanin content in transgenic lines and WT control, b: Cyanidin content, c: Pelargonidin content. Data were calculated from at least four biological repetitions and with error bars representing standard deviation. Statistical significance *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Gene expression studies
The expression level of poinsettia F3′H and the Cas9 gene, in the transgenic plants and the WT ‘Christmas Eve’ was determined by quantitative real-time PCR using actin and EF1A for normalization (primer sequences Suppl. Table S1). In general, the expression level of poinsettia F3′H for all transgenic lines was on a comparable level as in the WT, only for line B2.2 was it slightly lower (Fig. 7). We were able to detect Cas9 expression in all transgenic lines, whereas in the WT it was not observed. In all lines, expression of Cas9 between the propagated plants varied widely. The highest values were measured for line B2, whereas for the two other lines it was two or four times lower (Fig. 7). This correlates with the fact that a changed colouration could be observed only in line B2. In the other two lines (B158 and B284), the expression of Cas9 was probably not sufficient to perform the edit of the targeted gene.
Gene expression of poinsettia F3’H (left) and Cas9 (right) in transgenic poinsettia and WT. Quantitative expression was normalized to actin. Data were calculated from at least four biological repetitions and with error bars representing standard deviation. Statistical significance *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
F3’H cloning and heterologous expression in yeast
After PCR amplification of full size F3’H from transgenic plants from lines B2.1 and B2.2, two bands were detected (Fig. 8). One band with the original size present in the wild type around 1.5 kb, and second which was approx. 100 base pairs smaller. The same result was observed during screening, when also two bands were obtained when F3′H fragment was amplified as endogenous control (Fig. 3a, Fig. 4a). Therefore, F3′Hs from all transgenic plants and the WT were isolated and heterologously expressed in yeast in order to check the recombinant proteins for functional activity.
From the line B2, three different versions of F3′H cDNA clones were isolated, version 1 (v1) with a sequence identical to that of the WT, version 2 (v2) showing an insertion of a T in position 171, 3 nucleotides upstream PAM sequence, and version 3 (v3) showing a deletion of 126 nucleotides (Fig. 9, Suppl. Fig S1). F3′Hs clones isolated from lines B158 and B284 were identical to those of the WT (Suppl. Fig. S1, S2). In F3′H v2, the insertion of an additional T caused a shift in the open reading frame and thus, a premature termination of protein synthesis, whereas the deletion in F3′H v3, results in the loss of 42 amino acids between positions 37 and 79 (Suppl. Fig. S2). However, recombinant F3′Hs produced from F3′H v2 and v3 were not functionally active, which was tested with two main substrates of F3′H. All other recombinant F3′Hs were active, and the conversion of the substrate was on a similar level as that of the F3′Hs present in the WT (Table 4).
Discussion
Over more than 200 years of cultivation, the appearance of poinsettia has changed tremendously. Wild poinsettias are shrubs or small trees of up to 3 m height, with long internodes, a few stems, and narrow leaves and bracts (Trejo et al. 2012; Lee 2000). Commercial poinsettias as we know them from flower shops and supermarkets, in contrast, are small compact potted plants with multiple branches and wide large bracts (Trejo et al. 2012). The development of modern poinsettia took place in the twentieth century and the main milestones were the introduction of photoperiodic control, branching induction by transfer of phytoplasma via grafting, and the use of growth regulators (Taylor et al. 2011).
The colour spectrum of the bracts has also changed over time. Wild poinsettias are red or in a few cases white, but nowadays poinsettia are available in many different colour variations (Taylor et al. 2011), which gain more and more popularity (Barrett 2005). Apart from crossbreeding, radiation mutation is one of the basic methods to obtain novel bract colours (Broertjes and Harten 2013). The main drawback of this method is that mutations occur randomly, and a large number of plants has to be screened for interesting novel phenotypes (Kishi-Kaboshi et al. 2018), although in poinsettia the mutation rates leading to changes in the flower colour is relatively high (Lee et al. 2010). Genome editing methods like the CRISPR/Cas9 system offer significant improvement, as these allow precise targeting of the gene to be mutated (Liu et al. 2017). An additional advantage of CRISPR/Cas9 is that the transgene can be segregated in the progeny, so that a mutated plant without presence of a transgene can be obtained (Gao et al. 2016). CRISPR/Cas9 constructs are usually transferred to the plant cells by Agrobacterium- mediated transformation or particle bombardment (Erpen-Dalla Corte et al. 2019), which is a challenging process in some plant species.
Up to now, three classical poinsettia transformations mediated by Agrobacterium (Clarke et al. 2008) (Islam et al. 2013) (Sagvaag 2015) have been reported. To our knowledge, our approach is the first attempt to combine Agrobacterium- mediated transformations with CRISPR/Cas9 application in poinsettias. The approach faced problems with explant contamination due to insufficient surface sterilization. Clarke et al. (2008) suggested that the latex sap occurring at poinsettia cutting spots makes surface sterilization more challenging. The regeneration process of the chosen cultivar Christmas Eve was satisfactory. The percentage of somatic embryo producing explants was, however, lower than reported by Clarke et al. (2008) for cv. Millennium and is probably strongly dependent on the cultivar. The total transformation frequency during the transformation of cv. Millennium (Clarke et al., 2008) was similar to the value obtained in this study and it is around 2% (considering only explants that were not lost due to contamination).
Despite the fact that the transgene was detected in 14 regenerated plants on the genomic DNA level, only 3 of them showed transgene expression (6 were lost during the cultivation process). Of these, just one line (B2) developed bracts with brighter, reddish orange colour, than the WT (Fig. 5). When plants were propagated from the original shoots of B2, chimeric types were obtained in addition to the fully bright reddish orange phenotype. The chimeric plants can be described as non-pattern sectorial chimeras. This kind of mosaic phenotype is unstable (Frank and Chitwood 2016) and during further propagation, plants with different amounts of brighter areas were obtained. The occurrence of chimeric phenotypes was also reported for Japanese morning glory, in which DFR had been knocked out with the application of CRISPR/Cas9 system (Watanabe et al. 2017), and torenia where F3H was silenced (Nishihara et al. 2018). Probably chimeric tissue is a result of ongoing genome editing in line B2, despite that most of the genome edition events is considered to take place during early transformation stage (Nishihara et al. 2018). Only possibility to stop this process is crossing out Cas9, what is a problem in vegetatively propagated plants like poinsettia (Taylor et al. 2011). Chimerism of line B2 was confirmed by NGS data, when WT reads were detected together with mutated sequence (+ 1 nucleotide) in both fully reddish orange and the chimeric plant and by cloning of three versions of F3’H which were expressed in line B2. Lines B158 and B284 produced bracts with the same colour as the WT, indicating that the transgene could have been silenced. NGS data showed that only WT type reads are present in both lines indicating that the edition did not occur in those lines. This can happen, when the transgene is incorporated in a highly methylated region of the genome, or when more than one copy of the transgene is integrated (Vaucheret et al. 1998). It can also be explained by a transgene inactivation mechanism, in which the plant inactivates the transgene if recognized as a foreign DNA (Finnegan and McElroy 1994), which could happen before the edit occurred and as a result the plants without the edit and with no transgene expression are obtained.
The total amount of anthocyanins in the reddish orange plants obtained from line B2.1 was significantly lower than that in the WT. This is in agreement with the lower anthocyanin content of reddish orange compared with dark red cultivars (Nitarska et al. 2018). The level of pelargonidin remained almost unchanged in the transgenic plants compared with the wild type, only in line B2 was it significantly higher. Simultaneously, line B2.1 has a significantly lower cyanidin level, which indicates reduced F3’H activity compared with the WT (Fig. 6). The presence of cyanidin type anthocyanidins in line B2, however, suggests that the plant is heterozygous and that an allele with unmutated F3′H is also expressed. The lower anthocyanin content of the reddish orange line and the unchanged pelargonidin content can be explained by the low substrate specificity of the dihydroflavonol 4-reductase of poinsettia for DHK (Nitarska et al. 2018), which creates a bottleneck resulting in a slower anthocyanin formation and, therefore, in lower total anthocyanin concentrations, but because of the reduced F3′H activity an increased ratio of pelargonidin:cyanidin.
As reported for other plants (Jang et al. 2016), the expression of poinsettia F3′H was not affected by the CRISPR/Cas9 system. The expression level of Cas9 was much higher in the plants obtained from line B2 compared with plants obtained from lines B158 and B284, which correlates with the mutation efficiency. A positive correlation between Cas9 expression and mutation frequency was also reported in rice (Oryza sativa) (Mikami et al. 2015; Jang et al. 2016).
Cloning of F3’H from transgenic plants, expression in yeast and enzyme assay revealed that in line B2 at least three versions of F3′H are expressed. Version 1, with a sequence identical to the WT, showed no change in protein activity, which explains the presence of cyanidin in the bracts of line B2. Version 2, with insertion of T in position 171, which causes a shift in open reading frame and premature protein termination. The encoded protein was not active, which also supports the hypothesis that the mutation in the F3′H gene is a cause of change in poinsettia bract colour from red to orange (Nitarska et al. 2018). There is also a third version of F3′H, where 42 amino acids from the N-terminal of the protein were deleted. The obtained protein is almost not active, with only very slight activity with naringenin being observed. This version of F3′H was not detected during our NGS studies, probably due to big difference in the size, leading to this fragment being lost during the size selection process.
The CRISPR/Cas9 system is without doubt an interesting alternative for targeted mutagenesis in poinsettia. It should, however, be also taken into account that poinsettia transformation and regeneration is a challenging process and protocol optimization for each variety seems to be essential. Future work will concentrate on self-pollination of line B2 in order to obtain homozygous plants with more intense orange colour, which will accumulate prevalently pelargonidin. We are also planning to try to increase mutation efficiency in lines B158 and B284 by heat stress treatment as was performed by (LeBlanc et al. 2018).
Conclusion
In this study we reported for the first time successful targeted mutagenesis with the CRISPR/Cas9 system in the commercially important poinsettias. Poinsettias are propagated vegetatively, and selected varieties are frequently marketed as a series with a range of colours. This study puts forward CRISPR/Cas9 as an alternative to the process of irradiated mutation for breeding of colour variation, which is currently the standard practice in commercial poinsettia breeding. We have shown by way of F3′H as an example, that genome editing in poinsettia allows well directed knock-out of specific genes. With increasing knowledge on poinsettia metabolism, targeted knock-in or alteration of specific genes is only a question of time and would be strongly facilitated by the availability of a poinsettia genome. This includes another promising method in the repertoire of ornamental breeding and could be of relevance for other cultures where radiation breeding is frequently used, e.g. chrysanthemum, begonia or dahlia (Anne and Lim 2020). Genome editing methods will become fully exploitable once their status under EU law becomes uncoupled from that of heretofore conventional genetic engineering methods. In case that the EU regulation of genome edited plants will persist over the next decade, the knowledge obtained in this study could also be used to created new colour variants by conventional mutation techniques as e.g. Tilling. To the flavonoid community, this demonstrated that pelargonidin based flower colour can be achieved in an appropriate biochemical background even if DFR shows a low specificity for dihydrokaempferol, as previously hypothesized on the occasion of the escaped orange genetically modified petunia.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file. Primary datasets are available from the corresponding author on reasonable request.
Change history
02 October 2021
The original version of this article has been revised: The Funding section has been corrected.
References
Anne S, Lim JH (2020) Mutation breeding using gamma irradiation in the development of ornamental plants: a review. Flower Res. J. 28 (3):102–115
Barrett J (2005) Consumer poinsettia preference. L Gard Retail 4(2):1–4
Broertjes C, Van Harten A (2013) Applied mutation breeding for vegetatively propagated crops. Elsevier, Amsterdam
Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Biol 49(1):311–343
Chiruvella KK, Liang Z, Wilson TE (2013) Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5(5):a012757
Clarke JL, Spetz C, Haugslien S, Xing S, Dees MW, Moe R, Blystad D-R (2008) Agrobacterium tumefaciens-mediated transformation of poinsettia, Euphorbia pulcherrima, with virus-derived hairpin RNA constructs confers resistance to Poinsettia mosaic virus. Plant Cell Rep 27(6):1027–1038. https://doi.org/10.1007/s00299-008-0526-9
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, Pinello L (2019) CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37(3):224–226. https://doi.org/10.1038/s41587-019-0032-3
Ecke P (2004) The Ecke poinsettia manual. Ball Pub, Batavia
Erpen-Dalla Corte L, Mahmoud LM, Moraes TS, Mou Z, Grosser JW, Dutt M (2019) Development of Improved fruit, vegetable, and ornamental crops using the CRISPR/Cas9 genome editing technique. Plants (Basel, Switzerland) 8(12):601
Finnegan J, McElroy D (1994) Transgene inactivation: plants fight back! Nat Biotechnol 12(9):883–888
Frank MH, Chitwood DH (2016) Plant chimeras: the good, the bad, and the ‘Bizzaria.’ Dev Biol 419(1):41–53
Gao X, Chen J, Dai X, Zhang D, Zhao Y (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171(3):1794–1800
Halbwirth H (2010) The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. Int J Mol Sci 11(2):595–621
Halbwirth H, Kahl S, Jäger W, Reznicek G, Forkmann G, Stich K (2006) Synthesis of (14C)-labeled 5-deoxyflavonoids and their application in the study of dihydroflavonol/leucoanthocyanidin interconversion by dihydroflavonol 4-reductase. Plant Sci 170(3):587–595
Haselmair-Gosch C, Miosic S, Nitarska D, Roth BL, Walliser B, Paltram R, Lucaciu RC, Eidenberger L, Rattei T, Olbricht K (2018) Great cause—small effect: undeclared genetically engineered orange petunias harbor an inefficient dihydroflavonol 4-reductase. Front Plant Sci 9:149
Islam MA, Lütken H, Haugslien S, Blystad D-R, Torre S, Rolcik J, Rasmussen SK, Olsen JE, Clarke JL (2013) Overexpression of the AtSHI gene in poinsettia, Euphorbia pulcherrima, results in compact plants. PLoS ONE 8(1):e53377
Jang G, Lee S, Um TY, Chang SH, Lee HY, Chung PJ, Kim J-K, Do Choi Y (2016) Genetic chimerism of CRISPR/Cas9-mediated rice mutants. Plant Biotechnol Rep 10(6):425–435
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Kishi-Kaboshi M, Aida R, Sasaki K (2017) Generation of gene-edited Chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol 58(2):216–226
Kishi-Kaboshi M, Aida R, Sasaki K (2018) Genome engineering in ornamental plants: current status and future prospects. Plant Physiol Biochem 131:47–52
Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44(W1):W272–W276
LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93(2):377–386
Lee I-M (2000) Phytoplasma casts a magic spell that turns the fair poinsettia into a Christmas showpiece. Plant Health Prog 1(1):16
Lee EK, Kim WH, Kim ST, Kang SY (2010) Rooting, growth, and color mutation of poinsettias affected by gamma radiation. J Radiat Ind 4(3):253–257
Liu X, Wu S, Xu J, Sui C, Wei J (2017) Application of CRISPR/Cas9 in plant biology. Acta Pharmaceutica Sinica B 7(3):292–302
Ma X, Zhu Q, Chen Y, Liu Y-G (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant 9(7):961–974
Mikami M, Toki S, Endo M (2015) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34(10):1807–1815
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42(W1):W401–W407
Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M (2007) Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep 26(11):1951–1959
Nishihara M, Higuchi A, Watanabe A, Tasaki K (2018) Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol 18(1):1–9
Nitarska D, Stefanini C, Haselmair-Gosch C, Miosic S, Walliser B, Mikulic-Petkovsek M, Regos I, Slatnar A, Debener T, Terefe-Ayana D (2018) The rare orange-red colored Euphorbia pulcherrima cultivar ‘Harvest Orange’shows a nonsense mutation in a flavonoid 3’-hydroxylase allele expressed in the bracts. BMC Plant Biol 18(1):216
Pfaffl MW (2004) Quantification strategies in real-time PCR. AZ Quant PCR 1:89–113
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56(409):1–14
Sagvaag AK (2015) Genetic transformation of Poinsettia (Euphorbia pulcherrima). Norwegian University of Life Sciences, Ås
Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54(1):629–667
Schwinn K, Miosic S, Davies K, Thill J, Gotame TP, Stich K, Halbwirth H (2014) The B-ring hydroxylation pattern of anthocyanins can be determined through activity of the flavonoid 3′-hydroxylase on leucoanthocyanidins. Planta 240(5):1003–1010
Slatnar A, Mikulic-Petkovsek M, Veberic R, Stampar F, Schmitzer V (2013) Anthocyanin and chlorophyll content during poinsettia bract development. Sci Hortic 150:142–145
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84(6):1295–1305
Stewart R, Asen S, Massie D, Norris K (1979) The identification of Poinsettia cultivars by HPLC analysis of their anthocyanin content. Biochem Syst Ecol 7(4):281–287
Tanaka Y (2006) Flower colour and cytochromes P450. Phytochem Rev 5(2–3):283–291
Tasaki K, Higuchi A, Watanabe A, Sasaki N, Nishihara M (2019) Effects of knocking out three anthocyanin modification genes on the blue pigmentation of gentian flowers. Sci Rep 9(1):1–10
Taylor JM, Lopez RG, Currey CJ, Janick J (2011) The poinsettia: History and transformation. Chronica Horticulturae 51(3):23–28
Tong CG, Wu FH, Yuan YH, Chen YR, Lin CS (2020) High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotechnol J 18(4):889–891
Trejo L, Feria Arroyo TP, Olsen KM, Eguiarte LE, Arroyo B, Gruhn JA, Olson ME (2012) Poinsettia’s wild ancestor in the Mexican dry tropics: historical, genetic, and environmental evidence. Am J Bot 99(7):1146–1157
Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhettes S (1998) Transgene-induced gene silencing in plants. Plant J 16(6):651–659
Watanabe K, Kobayashi A, Endo M, Sage-Ono K, Toki S, Ono M (2017) CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci Rep 7(1):1–9
Yan R, Wang Z, Ren Y, Li H, Liu N, Sun H (2019) Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int J Mol Sci 20(12):2920
Zhang L, He L-L, Fu Q-T, Xu Z-F (2013) Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int J Mol Sci 14(12):24338–24354
Zhang B, Yang X, Yang C, Li M, Guo Y (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in Petunia. Sci Rep 6:20315
Acknowledgements
We would like to thank Christopher Schlosser for critical reading of the manuscript, Renate Paltram for technical support in HPLC analysis and Gabriel Alexander Vignolle for support during performing NGS analysis. The authors acknowledge Astrid Mach Aigner (Research Group Synthetic Biology and Molecular Biotechnology at the Institute of Chemical, Environmental and Bioscience Engineering, TU Wien) for making available the Illumina MiSeq system used in this study.
Funding
Open access funding provided by TU Wien (TUW). Open access funding provided by Austrian Science Fund (FWF). This project gratefully received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 675657 Flower Power and the Austrian Science Fund (FWF) P32901–B. The funding bodies had no role in the design of the study, collection, analysis or interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
DN, RB, TD and HH conceived the research. DN and RB designed the experiments. DN performed the experiments. RCL analysed the sequencing data. DN and HH wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Joyce Van Eck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nitarska, D., Boehm, R., Debener, T. et al. First genome edited poinsettias: targeted mutagenesis of flavonoid 3′-hydroxylase using CRISPR/Cas9 results in a colour shift. Plant Cell Tiss Organ Cult 147, 49–60 (2021). https://doi.org/10.1007/s11240-021-02103-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02103-5