Abstract
Somaclonal variation during in vitro culture is often an undesirable phenomenon but may also be a source of genetic variation useful for breeders. The molecular mechanisms underlying this phenomenon remain uncertain. In this study, we analyzed the fruit transcriptome of three independent cucumber lines using RNA-sequencing technology and performed an extensive in silico study to determine how somaclonal variation altered gene expression. Comparison of the transcriptome profiles with the wild-type cucumber ‘Borszczagowski B10’ revealed 418, 364, and 273 genes that were differentially regulated. We performed bioinformatic functional analysis, gene ontology classification, molecular network analysis, and explored differentially expressed genes associated with processes such as protein and nucleic acid binding, enzyme activity, signaling, transport, sugar and lipid metabolism. We determined that the differential gene expression may be caused by polymorphism in the genic region and may also be a result of interaction among molecular networks, which triggers specific pathways.
Key message
The somaclonal variation is the effect of the environmental impact on the physico-chemical state of the cell and DNA changes that influence gene expression and affect molecular networks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somaclonal variation is a phenomenon observed in cell and tissue cultures in vitro and was first described by Larkin and Scowcroft (1981). Somaclones show genetic changes associated with DNA sequences or epigenetic changes, but regardless of the genetic basis of the change, phenotypic variation occurs, which distinguishes the somaclones from the original parent (Guo et al. 2007). These changes may be temporary but may also be inherited by subsequent generations. Some genotypes are more susceptible to changes when cultured in vitro (Acquaah 2012). Somaclonal variation is often associated with the duration of in vitro culture of the explant (Hao and Deng 2002; Jevremović et al. 2012; Khan et al. 2011). Composition of the culture medium, in particular the presence of phytohormones, such as auxins, cytokinins, and sugar, can promote this phenomenon (Acquaah 2012; Cassels and Curry 2001; Smulders and de Klerk 2011). Furthermore, mechanical factors, such as damage to the explant or exposure to sterilizing agents, as well as disturbances in humidity, lighting, and temperature that affect plant transpiration can cause somaclonal variation (Cassells and Curry 2001; Krishna et al. 2016). The nature and frequency of changes are also determined by the explant itself, in particular its origin (Krishna et al. 2016). The molecular mechanisms leading to the formation of somaclonal variation still require clarification.
Somaclonal mutants can have potentially useful characteristics and generate variability in a population valuable for breeding. Somaclonal variation has become a useful tool in plant improvement, with regard to the utility of cultivars (e.g., yield, resistance to abiotic stress: unfavorable soil pH, excessive salinity, or drought) (Krishna et al. 2016), aesthetic qualities (e.g., traits of flowering and ornamental plants), and for scientific purposes (e.g., mutants that show modified expression of mitochondrial genes provide information on the function of such genes) (Bartoszewski et al. 2007).
Somaclonal variability allows the generation of plants that differ in phenotype (i.e., possess novel phenotypic traits) in a significantly shorter time and with reduced workload than with a classical breeding scheme, which typically extends from 10 to 15 years and is labor-intensive. This timeframe is due to the need to grow, evaluate, and cross several generations of individuals (Krishna et al. 2016). Additional advantages of using in vitro cultures as a source of variation include the notably lower cost compared with other methods and application of in vitro culture in breeding of plant species in which protoplast fusion or transformation is not possible (Karp 1995). Mutations that arise during in vitro culture can affect primary metabolism, but cells with such mutations are likely unable to survive the stressful culture conditions. Therefore, cells carrying mutations of genes responsible for secondary metabolism are indirectly selected (Evans 1989).
Despite many advantages, somaclonal variation has been underutilized as a tool for breeders, predominantly because of the randomness of the obtained phenotypes. Efforts are being made to understand the essence of somaclonal variation caused by the in vitro culture conditions. However, the phenotype of obtained regenerants is unpredictable and valuable organisms may be obtained during the first experiment or may fail to arise even after many attempts (Karp 1995). Moreover, some features are unstable and disappear after crossing or self-pollination (Karp 1995).
Regarding the study of cucumber (Cucumis sativus) somaclonal variation, the first experiments aimed to investigate the relationship between the regeneration system and genetic variation showed that the speed and nature of somaclonal changes strongly depends on the regeneration system and parameters such as culture time and composition of the selected medium (Pląder et al. 1998; Ładyżyński et al. 2002).
In this study we used three somaclonal lines (S1, S2, and S3) that differed phenotypically from the cultivar ‘Borszczagowski B10’ (the progenitor line) and were obtained by different regeneration methods (see details in the Materials and Methods section). The S1 line showed a mosaic phenotype (the combination of small yellow and irregular large silvery spots on the leaves), which is transmitted paternally and is considered to be associated with changes in the mitochondrial genome (Lilly et al. 2001). The S2 line showed an altered fruits phenotype, which are light green, glossy, without a waxy coat, and lack typical wards and netting. The S3 line produced shoot apices yellow-green in color. The phenotypic differences in the somaclonal lines are predominantly associated with the constituent factors and explants from which the culture was initiated (Skarzyńska et al. 2020).
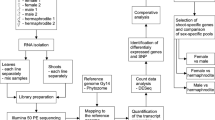
We sequenced the transcriptomes from fruit of the three somaclonal lines and the wild-type B10 line to determine the degree of variation among the transcriptomes. To evaluate how somaclonal variation influences the fruit transcriptome profiles, as they are marketable yield, we performed a comparative RNA-sequencing (RNA-seq) analysis and validation of differentially expressed genes (DEGs) in somaclones in relation to the wild type. The aims were, first, to determine if somaclonal variation phenomena could act as an active element and change the transcriptome profile. Second, we sought to detect common features for DEGs among the somaclonal lines to assess if this variation has a common factor, and third to identify genes that may be associated with the different phenotypic traits. By conducting a comparative analysis of the three somaclonal lines, we assessed the outcome of changed expression of genes and whether the changes were common to independent somaclones. In addition, we determined the chromosomal location of the genes to assess whether a specific genome region is more frequently subjected to rearrangements or whether the changes are randomly distributed across the genome. Based on findings from a previous comparative genomics study (Skarzyńska et al. 2020), we also determined genes that may be affected by structural polymorphisms, and how genomic variants influence gene expression and protein–protein molecular networks in the somaclones.
Materials and methods
Plant material and cultivation
A unique collection of cucumber somaclonal lines was used in this study. The somaclones S1, S2, and S3 possess the same genetic background as they were derived from the highly inbred homozygous cucumber ‘Borszczagowski B10’ during different in vitro culture procedures. Each somaclone was propagated by at least 10 self-pollinations and directed selection to maintain the specific phenotype. The S1 line was obtained by direct leaf regeneration (Malepszy et al. 1996; Bartoszewski et al 2004), the S2 line was obtained from leaf callus regeneration (Pląder et al. 1998), and the S3 line was obtained from cytokinin-dependent embryogenic suspension culture (Ładyżyński et al 2002). Plants were cultivated in the field from June to July 2014 and the mean temperature during the growing season was 18 °C. After self-pollination, tissues from 7-day-old fruit were collected, frozen in liquid nitrogen, and stored at − 80 °C. Three biological replicates were used for RNA-seq and real-time quantitative PCR (RT-qPCR) validation.
Isolation of RNA
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), with an additional step of DNase I treatment, in accordance with the manufacturer’s protocol. The nucleic acid concentration and quality were assessed with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For RNA quality assessment, the RNA integrity number (RIN) was calculated using a Bioanalyzer 2100 (RNA 6000 Nano Kit) in accordance with the manufacturer’s instructions (Agilent, Palo Alto, CA, USA). The RNA samples for sequencing contained total amounts of RNA ≥ 20 µg, RNA concentration ≥ 500 ng/µl, major ribosomal subunit ratio 28S:18S ≥ 1, and RIN ≥ 8. For RT-qPCR analysis, cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
Library construction and deep sequencing
To prepare the RNA-seq library, 10 μg total RNA was used per sample. Polyadenylated RNA purification, RNA fragmentation, cDNA synthesis, and PCR amplification were performed with the Illumina mRNA-seq Sample Prep Kit (Cat # RS-100–0801, Illumina, Inc., San Diego, CA, USA) in accordance with the manufacturer’s protocol. Parallel sequencing was performed using an Illumina HiSeq platform at the McGill University and Génome Québec Innovation Centre (Montreal, Canada).
Three replicates for each somaclonal line and three samples for the control B10 line were analyzed. Paired-end read sequences of 100 bp were generated. Read quality was evaluated based on the Illumina purity filter, percentage of low-quality reads, and distribution of Phred-like scores at each cycle. To assess the quality of short reads we used the FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) filter. Sequences are available at the National Center for Biotechnology Information Sequence Read Archive (BioProjects 578634 and 578623).
RNA-seq data analysis
To analyze gene expression and identify DEGs in somaclonal lines and B10 control plants, we performed transcriptome assembly with additional data from PCC Genomics (Osipowski et al. 2020). As a reference genome we used the cucumber genomic sequence B10v3 (GenBank: LKUO00000000). Gene expression values were estimated using Salmon (Patro et al. 2017) with sequence-specific and GC content bias enabled. Using the Limma package we performed differential expression analysis following the method of Law et al. (2016). As a second method, we performed DESeq pipeline calculation (Anders and Huber 2010). Genes were considered to be differentially expressed based on a false discovery rate (adjusted p value) < 0.001 and fold change > 1.5. To assess the reproducibility and repeatability of the probes and to cluster genes using Pearson correlation analysis, we used MeV 4.9.0 (http://mev.tm4.org). The principal component analysis was performed using R package with “ggplot2” package. Identified DEGs were compared with previously obtained genome sequencing data for somaclonal lines (BioProject PRJNA563814) (Skarzyńska et al. 2020) to locate single-nucleotide variations (SNVs) within the gene structure or upstream region (1500 bp upstream from the start codon). To visualize the DEGs position across chromosome maps, MapChart software (Voorrips 2002) and the R package “circlize” (Gu et al. 2014) were applied.
Validation of expression profiles of DEGs by qPCR
To verify the accuracy of DEGs identification we performed qPCR analysis for nine genes for the S1 line, 16 for the S2 line, and 17 for the S3 line. Using Primer3 (version 2.3.6) we designed primers for randomly chosen genes (Table S1). As an internal reference we used UBIep and CACS based on previously performed experiments by Skarzyńska et al. (2016). For each qPCR reaction we used three technical and three biological replicates. The qPCR assays were completed with 4 µL cDNA, the Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific). The qPCR program was 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C, using an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific). A melting curve analysis was completed immediately after the qPCR. The amplification efficiency for all primers was 74%–93%. The mean amplification efficiency was assessed with LinRegPCR (version 2015.3) (Ramakers et al. 2003). Relative expression levels were determined using the 2−ΔΔCt method with Rstudio and EasyqPCR from the Bioconductor software package (Pape 2012).
Functional annotation, gene ontology classification, and molecular networks
Blast2Go software was used for functional gene annotation and gene ontology (GO) assignment (Conesa et al. 2005) (Table S2). The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was accessed to estimate cis-acting elements and the possible functional influence of transcription factors (TF). The STRING algorithm (version 10.5) (Szklarczyk et al. 2017), using Arabidopsis thaliana as a model, was applied for an additional analysis of the possible interactions between the selected proteins coded by DEGs and genes affected by SNVs (G_SNVs) (Table S3), which were previously selected (Skarzyńska et al. 2020).
Results
Identification of DEGs
The total number of reads after sequencing ranged between 44 and 57 million for the analyzed lines with mean duplication 29.68% and average Phred quality 36 (Table 1).
Comparison of the fruit transcriptome profiles of the S1, S2, and S3 lines with that of the wild-type B10 (Table S2) revealed 418, 364, and 273 genes, respectively, that were differentially regulated based on the applied thresholds (Fig. 1).
Volcano plots of differentially expressed genes (DEGs) from three cucumber somaclonal lines (S1, S2, and S3) versus the wild-type ‘Borszczagowski B10’ (B10) line. Red dots represent significant up-regulated results; green dots represents significant down-regulated results; black dots indicate non-significant results. (Color figure online)
The number of down-regulated genes was 143, 90, and 109, whereas the number of up-regulated genes was 275, 274, and 164, for the somaclonal lines S1, S2, and S3, respectively (Fig. 2). Three common genes were differentially regulated for all somaclonal lines, of which one was down-regulated (G1138) and two were up-regulated (G20095 and G20923). The expression of 66 genes was differentially regulated in two lines (Fig. 2, Table S2), comprising 20 down-regulated and 46 up-regulated genes. These groups of genes followed the same direction of differing expression in the two somaclonal lines.
The expression profiles of DEGs analyzed by hierarchical clustering (HCA) are shown in Fig. 3a. The probes were highly replicable in all four lines (B10, S1, S2, and S3). Hierarchical clustering also provided evidence that the sets of DEGs differed between the individual somaclonal lines, showing that each somaclone represents an independent somaclonal event (Fig. 3b–d). To examine the trends in expression changes among the sample groups, all DEGs were assigned to six clusters by the K-means method using MeV software. In this set of three somaclonal lines, the distinctness of each line with regard to the direction of gene expression was as follows: up- and down-regulated for the S1 line—clusters 1 and 2, for the S2 line—clusters 3 and 4, and for the S3 line—clusters 5 and 6, respectively. The up- or down-regulated DEGs were specific to individual groups of samples representing a single line (S1, S2, or S3), but the remainder of the genes were similar to the control B10 line. Nonlinear PCA for all genes (21661) detected in the cucumber reference genome B10v3 (Osipowski et al. 2020) revealed structure in the data, with obvious subgroups of the somaclonal lines, especially with regard to DEGs (Fig. 3e, f).
Expression profiles of differentially expressed genes (DEGs) in the fruits of three cucumber somaclonal lines (S1, S2, and S3) and the wild-type ‘Borszczagowski B10’ (B10) line. a Heat map for cluster analysis of the DEGs. The expression levels are indicated at the top of the heat map, with red and green indicating down-regulated and up-regulated expression, respectively. b–d K-means cluster analysis of genes that were up-regulated (left) and down-regulated (right) in the S1 line (b), the S2 line (c), and the S3 line (d). e Principal component analysis of all identified transcripts from the four analyzed lines. f Principal component analysis of DEGs from the four analyzed lines. (Color figure online)
Detection of polymorphism in the gene structure of DEGs
We performed a comparison of previously detected SNVs within the cucumber genome (Skarzyńska et al. 2020) with DEGs across the genome of the somaclonal lines. Taking into consideration the number of genes with SNVs across the gene structure (exon and intron) and upstream region (− 1500 bp), the highest number of DEGs were observed in the S3 line—99, whereas 37 and 38 were observed in the S1 and S2 lines, respectively (Table 2). The number of SNVs in the genic region were 193 in the S1 line, 95 in the S2 line, and 766 in S3 line. The number of SNVs in the promoter region was highest in the S3 line (382) compared with the number in the S1 line (84) and the S2 line (61).
Chromosomal distribution of DEGs
The DEGs were mapped to individual chromosomes based on previously published data regarding contig positions on chromosomes (Osipowski et al. 2020; Skarzyńska et al. 2020). Of the total DEGs, 92.11% (385), 90.66% (330), and 76.56% (209) were assigned to chromosomes for the S1, S2, and S3 lines, respectively. The highest number of DEGs were mapped to chromosomes 6 and 3 (Fig. 4, Table S4). The distribution of the identified DEGs on the cucumber chromosomes indicated that the genes were evenly distributed among the chromosome and the genes did not form clusters (Fig. 4, Table S2).
Distribution of differentially expressed genes (DEGs) of cucumber somaclonal lines S1, S2, and S3 on individual chromosomes. The number of DEGs is indicated as dashes and dots. A red dash represents up-regulated genes and a green dash represents down-regulated genes; a black dot represents protein-coding genes and a grey dot represents lincRNA. Order starting from the inner circle: chromosome coverage (contigs colored), S1, S2, and S3 line. (Color figure online)
Confirmation of Illumina RNA-seq expression by qPCR
To validate the results of the DEGs analysis based on RNA-seq data, 42 randomly selected DEGs were subjected to experimental verification by qPCR (Fig. 5, Table S1). Nine genes were selected for the S1 line, 16 genes for the S2 line, and 17 genes for the S3 line. The qPCR and RNA-seq data were compared (Fig. 5). Highly reliable reference genes were used to normalize the qPCR data using the 2−ΔΔCt method. The qPCR and RNA-seq data for the analyzed genes were positively correlated for all analyzed DEGs except one (G1468).
Functional categories of DEGs
Genes subject to differential regulation were annotated using BLAST and categorized in accordance with GO terms using Blast2Go software. In addition, the genes were classified with a set of plant-specific GO slims and were clustered into three main categories: biological process, molecular function, and cellular component (Fig. 6). Of the total genes, 307 genes from the S1 line, 238 from the S2 line, and 130 from the S3 line were assigned to at least one category. Among these genes, for the S1, S2, and S3 lines, 178, 141, and 75 were assigned to the biological process category; 258, 184, and 106 were classified to cellular components; and 181, 145, and 82 were assigned to molecular function, respectively. The number of GO terms assigned in individual categories were as follows: 682, 477, and 322 for biological process, 1133, 683, and 429 for cellular component, and 309, 233, and 151 for molecular function for the S1, S2, and S3 lines, respectively. Detailed analysis of GO slim assignments are presented in Table S5.
The most abundant terms according to GO slim terminology in the biological process category were cellular processes (16.28%, 13.42%, and 14.91%) and metabolic processes (12.02%, 10.90%, and 14.91%) for the S1, S2, and S3 lines, respectively. The terms response to stress (7.04% and 5.90%) and biosynthetic processes (5.43% and 5.90%) were the most frequent for the S1 and S3 lines, respectively. Transport (5.13%) and response to abiotic stimuli (5.13%) were next most frequent terms for the S1 line. The group multicellular organism development was abundant in the S2 (7.97%) and S3 (5.28%) lines. The post-embryonic group and response for abiotic stimuli were also frequent (5.45% and 5.24%, respectively) in the S2 line.
In the molecular function category, the most frequent terms were for genes encoding proteins connected with catalytic activity (18.12%, 16.74%, and 25.83%) and protein binding (14.56%, 16.74%, and 13.25%) in the S1, S2, and S3 lines, respectively. The next most frequent groups were binding (12.30%) and transporter activity (9.06%) for the S1 line, transcription factor (16.31%) and DNA binding (10.30%) for the S2 line, and binding (12.58%) and transferase activity (9.27%) for the S3 line.
With regard to the cellular component category, the most abundant GO terms were connected to vacuole, thylakoid (17.12%, 16.11%, and 16.55%), ribosome (14.3%, 9.96%, and 12.59%), and extracellular matrix (11.3%, 12.59%, and 11.42%) in the S1, S2, and S3 lines, respectively. The next most numerous groups were peroxisome (7.76% and 5.36%) and plasma membrane (7.03% and 5.59%) for the S2 and S3 lines, respectively, and plastid (7.86%) and plasma membrane (5.59%) for the S1 line.
Bioinformatic analysis of upstream regions
Analysis of the promoters with the PlantCARE database detected 36 830, 31 923, and 24 107 motifs, and the average number of motifs per promoter region was 88 (range 47–266), 87 (51–194), and 88 (26–236) for the S1, S2, and S3 lines, respectively. The 12 most abundant motifs were common to the three lines analyzed. Less frequent motifs varied depending on the line (Fig. 7, Table S6).
The most numerous functional group of motifs across the promoter region was “core promoter and enhancer elements”, which comprised 16 055, 14 284, and 10 495 motifs in the S1, S2, and S3 lines, respectively (Table S6). The cis-acting elements without specific function were classified to the group “other”, which was also numerous in all somaclonal lines (S1: 2093, S2: 1833, and S3: 1377). The specific functional groups predicted for detected cis-acting elements are presented in Fig. 8. The most numerous group among those with an assigned function was “reaction to light” (S1: 2579, S2: 2290, and S3: 1630). The next most numerous groups were methyl jasmonate response, abscisic acid response, anaerobic induction, and others (Fig. 8).
In silico modeling of interaction of proteins encoded by DEGs
Molecular protein networks for each somaclonal line were constructed based on the results of a STRING analysis. For the STRING analysis we used as input DEGs that encode proteins and genes previously shown to contain SNVs (G_SNVs) (Skarzyńska et al. 2020). In total, we used 537, 454, and 388 identifiers and among these 418, 364, and 273 DEGs and 119, 90, and 115 G_SNVs for the S1, S2, and S3 lines, respectively. From the STRING database, 495, 392, and 294 proteins were annotated with average nodes degree 8.14, 1.47, and 1.03 for the S1, S2, and S3 lines, respectively.
In the S1 line, 152 proteins were isolated nodes and 343 proteins (188 up-regulated and 79 down-regulated encoded by DEGs, and 76 encoded by G_SNVs) were included in networks (Fig. 9a, Table S3). The most extensive network consisted of 336 nodes, among which 182 were coded by up-regulated and 74 by down-regulated DEGs and 74 were coded by G_SNVs. In addition, five smaller networks were identified: two consisting of four and three nodes (composed of proteins encoded by DEGs), and three two-component networks (involving proteins encoded by DEGs and G_SNVs). The highest number of proteins of specific function were connected with response to stimuli, catalytic activity, ion and cation binding, and with plastid and chloroplast cell compartment.
In silico analysis using STRING software of protein–protein interactions among proteins encoded by differentially expressed genes (DEGs) and genes influenced by small genomic changes (G_SNVs) of the cucumber somaclonal lines S1, S2, and S3. a the network in the S1 line, b the network in the S2 line, c the network in the S3 line. Isolated nodes were excluded. Red nodes represent proteins encoded by up-regulated DEGs, green nodes represent proteins encoded by down-regulated DEGs, and grey nodes represent proteins encoded by G_SNVs
The STRING analysis for the S2 line (Fig. 9b, Table S3) revealed 171 unconnected proteins and 221 proteins (127 up-regulated and 46 down-regulated encoded by DEGs, and 48 encoded by G_SNVs) were included in the networks. The expanded network consisted of 205 nodes in which 122 nodes were from up-regulated and 40 from down-regulated DEGs and 43 from G_SNVs. Networks consisting of five nodes, two four-component networks, three three-component networks, and seven two-component nets were observed. The smaller networks mostly consisted of proteins coded by DEGs but a number of five-node networks were coded also by G_SNVs. In the networks, the majority of proteins with a characterized function were associated with DNA binding TFs, DNA binding, and catalytic activity and membranes with regard to cellular components.
In the S3 line, 152 proteins were assigned as single and 142 proteins (61 up-regulated and 34 down-regulated encoded by DEGs, and 47 encoded by G_SNVs) were included in networks (Fig. 9c, Table S3). The networks in this line were not extensively branched as was observed for the S1 and S2 lines. The largest network consisted of 72 nodes among which 24 proteins were encoded by up-regulated DEGs, 16 by down-regulated DEGs, and 32 by G_SNVs. Several smaller networks were built, such as one nine-component network (consisting of nodes from DEGs), one seven-component network (only up-regulated DEGs and G_SNVs), one six-component network, two four-component networks, four three-component networks, and 14 two-component networks. In the smaller networks, nodes were observed in various groups of proteins coded by up- and down-regulated DEGs and by G_SNVs. In the networks the most characterized proteins were associated with catalytic and oxidoreductase activity, and cofactor and ion binding.
Discussion
The first study of somaclonal variability in cucumber investigated the relationship between the regeneration system and the resulting variability (Pląder et al. 1998). This aspect has been studied in detail by testing a variety of factors, such as regeneration methods, cultivation times, and modifications to the Murashige and Skoog medium (Ładyżyński et al. 2002). A recent comparative genome analysis of the S1, S2, and S3 somaclonal lines was performed to describe the differences at the molecular level using whole-genome sequencing technology (Skarzyńska et al. 2020).
In the present study, we analyzed the transcriptomes of cucumber fruit using RNA-seq technology and performed an extensive in silico study of the DEGs to investigate how the somaclonal variation altered the transcriptome profiles. The three somaclonal lines analyzed were obtained using different regeneration techniques from the same cultivar (‘Borszczagowski B10’). Comparison of the fruit transcriptome profiles of the S1, S2, and S3 line with the wild-type B10 (Table S2) revealed 418, 364, and 273 genes that were differentially regulated.
Reproducibility
The majority of DEGs showed a consistent expression trend among the three biological replicates, which was highlighted in the heatmaps (Fig. 3). This result indicated the field environment had minimal impact on growth of the replicates and that the experimental data are reproducible. The RNA-seq results were confirmed with high accuracy by qPCR analysis, which proved the high efficiency of the method used and the significant coverage and reliability of algorithms predicting the resulting transcripts.
Chromosomal location
The DEGs were distributed on each chromosome (Fig. 4), with no concentration on specific chromosomes, and no concentration on certain regions of chromosomes. No significant correlation was observed between the number of differential sequences on a given chromosome and its length, which indicated that the changes were entirely random.
Functionality of DEGs
The identified DEGs were generally unique for each somaclonal line, but three genes showed a similar changed pattern of expression in all somaclonal lines - one gene was down-regulated (G1138) and two were up-regulated (G20095 and G20923). The G1138 gene encodes a cell signaling peptide that may regulate plant stress, growth, and development. It mediates rapid alkalinization of extracellular space by mediating a transient increase in the cytoplasmic Ca(2+) concentration leading to calcium-dependent signaling events through a cell surface receptor and concomitant activation of certain intracellular mitogen-activated protein kinases. The G20923 gene encodes a CACTA transposon protein belonging to En/Spm subclass. The role of this protein is unclear, but it has been suggested that it could contribute regulatory sequences that may alter gene expression (Wicker et al. 2003). The function of the third gene, G20095, is unknown.
Genes that are common to the two lines are also noteworthy, especially those that have a similar expression pattern and undergo increased regulation (with high fold change) compared to the control line. One of such genes is G6620 (up-regulated in S1 and S2 lines) encoding ubiquitin-protein ligase. These proteins take part in the ubiquitination process, as a result of which proteins are marked for degradation. Ubiquitination regulates diverse areas such as cell trafficking, DNA repair, signaling and is of profound importance in cell biology (Teixeira and Reed 2013). Other genes: G9830 (up-regulated in S2 and S3 lines) encodes mate efflux protein function as fundamental transporters of metabolic and xenobiotic organic cations (Omote et al. 2006) and G143838 (up-regulated in S2 and S3 lines) encodes peroxidase which belong to large group of enzyme which generally break down peroxides and take part in stress tolerance (Bolwell et al. 2002). Highly expressed in both lines (up-regulated in S2 and S3 lines) is also gene G14398 that encodes protein of unknown function with DUF604 domain (domain of unknown function) which is a conserved region found in several uncharacterised plant proteins (Bateman et al. 2010).
Interestingly, even though the DEGs are predominantly line-specific (Fig. 3), the ontology functional groups were similar in all lines. We speculate that functional groups influence the physiological and chemical cell state and lead to the process of somaclonal variation. A similar conclusion was driven during comparative genome analysis (Skarzyńska et al. 2020) and is also described by Krishna et al. (2016).
It is possible that the altered expression of certain genes is not directly due to changes induced by passing through in vitro culture, but rather to the changed activity of the cell's transcriptional apparatus. This phenomenon makes these genes effectively indirect effects of somaclonal variation.
Protein binding and enzyme activity
Protein modification is a common regulatory mechanism among eukaryotic organisms involved in the regulation of many cellular and developmental processes at the post-translational level (Liu et al. 2013). These processes in plants include regulation of the cell cycle, tissue differentiation, response to phytohormones, protein transport (Fang et al. 2015), response to pathogenic microorganisms (Sharma et al. 2016), and TFs. The largest group of DEGs were those associated with enzymatic proteins (in total 33.33%, 29.18%, and 43.71% in the S1, S2, and S3 lines, respectively). Genes with an altered expression profile encoded proteins such as transferases, kinases, hydrolases, nucleases, and enzyme regulators, or regulators of structural molecule activity and activity of other enzymes (Fig. 6b, Table S5). This result indicated that enzyme activity may also be disrupted during in vitro culture compared with that under normal plant growth. A large group of DEGs was associated with protein binding (14.56%, 16.74%, and 13.25% for the S1, S2, and S3 lines, respectively), thus the majority of them interact selectively and non-covalently with other proteins or protein complexes (including other non-protein molecules). Higher enzymatic activity and protein binding can be an adaptive response, which can lead to far-reaching somaclonal changes resulting in a visible phenotypic effect. Taking into account biological processes, the majority of DEGs were involved in responses to various types of stimuli (in total 18.91%, 17.19%, and 16.15% in the S1, S2, and S3 lines, respectively), biosynthesis processes (5.43%, 3.56%, and 5.9%), and protein metabolism (2.64%, 1.05%, and 1.24%) (Fig. 6a, Table S5). Proteins encoded by DEGs were also involved in processes regarding cellular organization, development, and reproduction (in total 12.02%, 19.92%, and 15.53%) (Fig. 6a, Table S5).
It can be concluded that plants regulate enzyme activity in processes that respond to the in vitro culture conditions, which in turn influence the change in response processes to various factors, as well as cell growth, development, differentiation, and biosynthesis. Enzyme activity could also be modulated by a variety of stimuli, which may cause specific reactions to be switched on or off. In addition, enzymes can interact to influence pathways leading to somaclonal variation (Claaßen et al. 2019).
Receptors, signal transduction and transporters
The environment in which the organism is located may initiate the entire cascade of gene expression events leading to somaclonal variation. Receptors play an important role in receiving external signals and transmitting them inside the cell. Molecular functions associated with receptor activity and binding (0.97%, 0.86%, and 0.66% in the S1, S2, and S3 lines, respectively) and signal transduction and transport (11%, 5.58%, and 3.97%) were assigned to DEGs (Fig. 6b, Table S5). Regarding biological processes, these proteins participate in signal transduction, transport, and cell communication (10.85%, 8.18%, and 5.59%) (Fig. 6a, Table S5). These molecules enable communication inside the cell, between all organelles, or even between cells. Without transporters, the coordination of all cells would not be possible and complex pluricellular organisms could not exist. These proteins, as well as ions (e.g., Mg(2+), Fe(2+), and Zn(2+)), are also associated with phytohormone responses, cell differentiation, and distribution of other diverse substances within the cell.
Phytohormones such as auxins, cytokinins, brassinosteroids, abscisic acid, and jasmonic acid strictly regulate plant growth (Dewitte and Murray 2003). Somaclonal changes imply a disrupted hormonal signal transduction, which in turn results in changes in the cell cycle and growth, which are characteristic phenotypic features confirmed by slower growth in somaclones. Thus, if transport is disrupted, important changes in the plant can be expected, such as differential expression of certain phytohormone-related genes, and light- or stress-responsive genes. A promoter analysis may confirm this hypothesis because hormone-, light-, and stress-responsive factors are among the most abundant elements in the promoter region. Disturbances in transport can lead to uneven distribution of critical substances required for plant development, such as assimilates, ions, proteins, and other molecules, which in consequence may disrupt plant growth, chlorophyll synthesis, or metabolism of sugars and nucleic acids (Chinnusamy et al. 2004).
Nucleic acid binding and transcription factor activity
The other large groups of DEGs identified were associated with nucleic acid binding (20.06%, 18.03%, and 18.54 in the S1, S2, and S3 lines, respectively) and TFs (5.18%, 16.31%, and 5.96%) (Fig. 6b, Table S5). These genes are associated with transcription regulation at various levels activated at different times of the cell's life and their altered expression may result in a broad spectrum of dysfunctions in somaclonal lines. This group mainly consisted of genes that are directly responsible for transcription, such as RNA polymerase subunits or genes encoding polymerases or proteins that bind to DNA that act as TFs. The PLATZ TFs are implied to participate in cell division and can act as a repressor or as an activator (Kim et al. 2018). The HD-Zip TFs are predominantly involved in plant development (Ariel et al. 2007). Variable expressions of genes encoding PLATZ (in the S3 line) and HD (in the S2 line) TFs may explain the slower growth of these lines. The TFs containing a NAC domain are generally suggested to be involved in biotic and abiotic stress tolerance (Puranik et al. 2012).
The numerous genes with altered expression encoding TFs that play roles in reception of stimuli during stress included WRKY TFs, which can be differentially expressed in response to wounding (Eulgem et al. 2000), bHLH TFs, which show an extremely wide range of actions, including stress response (Feller et al. 2010), ERF TFs, which predominantly regulate stress responses (Xu et al. 2008), and Heat Shock TFs, which are involved in the expression of heat shock proteins in response to stresses (Lohmann et al. 2004). These TFs might reflect the stressful environment that the plants experience during in vitro culture. Heat Shock TFs might bind to a stress-responsive element located in the promoter region of DEGs of the S2 line. The bHLH TFs are involved in phytohormone signaling, including DOF and bZIP TFs (Feller et al. 2010; Noguero et al. 2013; Jakoby et al. 2002). The KAN TF is known to regulate auxin transport (Ilegems et al. 2010). In somaclonal lines, phytohormone-responsive elements were important cis-acting elements in the promoter region. In addition, bHLH and MYB TFs can interact in the flavonoid biosynthesis pathway (Feller et al. 2010). That might explain the presence of motifs involved in the regulation of several flavonoid biosynthesis genes in the promoter region of DEGs. Furthermore, DOF, bHLH, and bZIP TFs are involved in light signaling (Noguero et al. 2013; Jakoby et al. 2002; Feller et al. 2010).
Sugar and lipid metabolism
Among the biological processes in which proteins encoded by DEGs participate, attention should be paid to two relatively numerous groups: sugar metabolism (1.76%, 3.35%, and 1.86% in the S1, S2, and S3 lines, respectively) and lipid metabolism (1.31%, 2.31%, and 3.11%) (Fig. 6a, Table S5). Lipids are present in membranes and influence cytoskeleton plasticity. Plants require lipids for membrane biogenesis, as signal molecules, and as a form of stored carbon and energy (Schmid 2015). Saccharides play an important role in the plant's life: they are structural and storage substances, respiratory substrates, and intermediate metabolites of many biochemical processes. Changes in the concentration, qualitative composition, and transport of sugars occur continuously in plant tissues, during the day and night, as well as during subsequent developmental stages (Ciereszko 2018).
Binding
A large number of proteins were associated with binding (12.3%, 8.15%, and 12.58% in the S1, S2, and S3 lines, respectively), among which were proteins connected with management of metal ion binding. This result suggests the occurrence of disorders in the maintenance of metals homeostasis. This phenotype may be similar to that of plants with a deficiency or excess of such minerals. Certain DEGs affect the homeostasis of magnesium, iron, zinc, and calcium ions, which, in combination with a large number of proteins and pectins that bind these ions, can lead to their physiological deficiency. Deficiency of calcium ions results in growth retardation and leaf deformation, which coincides with the overall growth phenotype of the three analyzed somaclonal lines. Magnesium is a component of chlorophyll and is required for the activity of many enzymes, and thus is an essential element in plant cells (Guo et al. 2016). The primary symptom of magnesium deficiency is chlorosis, directly caused by termination of chlorophyll synthesis. An additional symptom is inhibition of plant growth (Guo et al. 2016). Iron and zinc deficiencies cause chlorosis by inhibiting the synthesis of chloroplast proteins and also result in growth disorders (Zhang et al. 2019). Zinc ions act as an activator of many enzymes and bind to the zinc finger protein domain. A deficiency of zinc ions leads to a number of disorders in the metabolism of nucleic acids and sugars, what was correlated with growth inhibition of plants (Zhang et al. 2019; De Assuncao et al. 2018). An important point is that, the protein coded by DEGs (pointed in this study) in the studied genes many have the ability to bind ATP. This observation suggests that there are changes in processes involving proteins that use ATP as a substrate, which may also cause changes in transmembrane transport or changes in osmotic pressure, what was also confirmed in others study (Clausen et al. 2017).
Promoters
The core promoter elements are essential for transcription because they include the start site of transcription. These elements can comprise motifs such as a TATA-box, which is the most important core promoter element (Burke and Kadonaga 1997). The CAAT-box is also considered to be a core promoter element (Lee-Huang et al. 1993) and influences the transcriptional initiation frequency (Kusnetsov et al. 1999). This explains their abundance among the analyzed promoter sequences in the present study, because the gene cannot be transcribed and expressed without the presence of a core promoter element. The other core promoter motifs, which were less abundant in the analyzed sequences, show greater gene specificity. The majority regulate gene expression in response to diverse factors, such as light, stress, or phytohormones. The MYB transcription factors (TF) are present in many eukaryotes and are described as regulatory proteins (positive or negative) regulating pathways involved in: primary and secondary metabolism, growth and development, as well as biotic and abiotic stress responses (Chen et al. 2019a; b). However, authors suggest that there are still many questions to be answered, like: "how MYB repressors perceive environmental signals, what is the role of micro-RNA in the regulation of MYB repressors, does post-translational modifications play any role in the regulation?" The MYB motif is known to regulate genes that are mostly specialized for cell proliferation and cell shape, in response to phytohormones such as gibberellins and abscisic acid (Martin and Paz-Ares 1997). Moreover, recently described results (Ding et al. 2020) characterize a R2R3-MYB activator and a R3-MYB repressor in monkeyflowers (Mimulus). They demonstrated that the properties of these two proteins correspond to an activator-inhibitor pair in a two-component, reaction–diffusion (RD) system, explaining the formation of dispersed anthocyanin spots in monkeyflower petals.
The MYC motif regulates genes that might be involved in the cell cycle, differentiation, and death (Amati and Land 1994). It is also implicated in certain phytohormone pathways, such as the jasmonic acid signaling pathway in response to abiotic stress (Lorenzo et al. 2004; Boter et al. 2004). Latest research (Chen et al. 2019b) demonstrate that the C-terminal region of some MYC genes from Triticum aestivum, Oryza sativa, and Brachypodium distachyon is conserved and includes the typical basic region and HLH domain, and most MYCs contain leucine zipper (bZIP) domain. Authors demonstrated that most MYC genes are tissue specific and expressed in roots, stems, leaves, and inflorescences. In combination with the identification of many cis-elements related to plant growth and development and hormone stress, these results further suggest their functions in growth/development, and in response to environmental stresses. MYC genes interact with MYB genes (Lorenzo et al. 2004). This may explain why both motifs were detected in approximately equivalent quantities among the analyzed sequences in the three somaclonal lines. The ABRE motif is involved in stress tolerance (Yoshida et al. 2010), and other motifs such as the G-box and B-box can regulate multiple genes depending on a wide range of factors (Menkens et al. 1995). Their broad range of possible actions explains their abundance in the present study.
The most abundant functional category was light-responsive elements. Plant life is conditioned by light as it is essential for photosynthesis, which enables the production of sugars for plant growth and development. Artificial light is used during in vitro culture, although the intensity and wavelength differ from those of sunlight. The daily photoperiod is identical, typically 16 h of day and 8 h of night (Comino et al. 2019), and the light does not increase and diminish gradually as the sun rises or sets. These artificial conditions suggest that the expression of genes regulated by light-responsive elements is altered and prolonged exposure to abnormal lighting may cause long-term adaptation of the plant through mutations. Indeed, a somaclone of Prunus avium L. ‘Hedelfinger’ shows a different response to the light compared with the wild type (Piagnani et al. 2002). It is likely that a similar process affecting light-responsive genes occurred in somaclonal lines. Another numerous group of motives found in the DEGs promoters are those that take part in the response to plant growth regulators. Phytohormones are required in small amounts but are critical for the functioning of plants on many levels of cell development and differentiation. Therefore, the presence of these motifs in the promoter of DEGs is of obvious importance.
Influence of genome changes on the transcriptome of somaclones
The somaclonal lines analyzed in the present study were derived from in vitro culture and self-pollinated under controlled greenhouse conditions (Skarzyńska et al. 2020). For the current experiment the lines were grown in the field. Previously, we also examined three independent transgenic cucumber lines (Pawełkowicz et al. 2020) that were derived from the same B10 line and, after transformation, were regenerated in in vitro cultures. The changes in gene expression between transgenic lines compared with the control line (plants were grown in the same field experiment as the somaclones) were as follows: three, six, and 38 DEGs in the respective analyzed transgenic lines (Pawełkowicz et al. 2020). The number of DEGs detected in the somaclonal lines was significantly higher than those observed among the transgenic lines. We presume that this variability is influenced, among others, by the composition of the medium and the maintanance time of plants in the in-vitro culture, which in the transformation experiments (Szwacka et al. 1996) were different than in the experiment in which these somaclonal lines were obtained. Therefore, the detected transcriptome changes in these somaclonal lines are due to the phenomenon of somaclonal variation and had to be established in these lines. Thus, the first step in the analysis of the somaclonal lines was the previously performed comparative genomic analysis (Skarzyńska et al. 2020). The analyzed polymorphisms of the S1, S2, and S3 lines were predominantly located in intergenic regions. Some 35%–41% of polymorphisms were positioned in genic regions (exons, introns, and untranslated regions) with the highest proportion located in introns (61%–67% of the variants located within genes) and a small percentage (1.08%, 0.96%, and 0.19% for the S1, S2, and S3 lines, respectively) had a strong effect on protein functionality (Skarzyńska et al. 2020).
Drawing on the findings of Skarzyńska et al. (2020), herein we identified the highest number of DEGs that showed changes in the genic structure and the promoter region in the S3 line, compared with those of the S1 and S2 lines, based on the number of DEGs with SNVs. We concluded that structural changes in the DNA of both genes and their promoters can affect their variable expression, and describe such DEGs as “primary”. It is likely that the remaining DEGs (without SNVs) are interdependent with primary DEGs as well as with other changes that have occurred in the genomes, in particular in other gene regions and probably in intergenic regions. Even a single, small change may affect the expression of many genes, especially in the "ubiquitous" highly active genes that play important roles in the cell. Genomic changes may result in the appearance of proteins with a different structure, which will likely be less efficient in fulfilling their functions, thereby affecting the expression of DEGs, which we can define as “secondary”. In the previous comparative genomic analysis (Skarzyńska et al. 2020), 179 genes were detected in which SNVs could impact on protein function. Among these genes, according to the GO classification, the most abundant groups with regard to molecular function were biosynthetic process, nucleobase-containing compound metabolic process, transport, and cellular protein modification process. We consider that the aforementioned functions are connected with other DEGs associated with the processes highlighted herein which justify our hypothesis (Fig. 10). Certainly, the changes in gene expression are caused not only by SNVs in the DNA structure, but are also the result of many additional factors, both physical and chemical, which influence the general state of the cell and activate transcription factors, which in turn regulate transcription. In addition, it has been shown that changes in DNA can also affect changes in the methylation pattern, which in turn contributes to changes in gene expression. Moreover, changes in methylation patterns in somaclones may appear not only in the R0 generation but may also be preserved in the following generations (Guo et al. 2007).
Conclusions
We conclude that the phenotype of the somaclonal lines is likely due to the interaction of proteins belonging to many functional groups. Functional analysis of DEGs revealed that the genes are involved in diverse processes such as protein modification, enzyme activity, transcription, signal transduction, transport, and ion homeostasis. The changes in phenotype are caused by the activation or repression of genes that in their promoter region contain elements associated with light response, methyl jasmonate response, abscisic acid response, anaerobic induction, and phytohormones. The genes with altered expression can be divided into two groups: “primary” and “secondary”, which are the result of changes in molecular network interaction across the “primary” genes. The DEGs are distributed on all chromosomes with no clear preference for the chromosomal region where the gene is located. No significant correlation was detected between the number of differential sequences on a given chromosome and its length, which indicates that the changes were random.
Data availability
All clean reads generated by Illumina sequencing in this study have been deposited in the National Center of Biotechnology Information Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra) under the BioProject accessions 578634 and 578623.
References
Acquaah G (2012) Clonal propagation and in vitro culture. In: Acquaah G (ed) Principles of plant genetics and breeding, 2nd edn, pp 146–170. https://doi.org/10.1002/9781118313718.ch8
Amati B, Land H (1994) Myc–Max–Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev 4:102–108. https://doi.org/10.1016/0959-437X(94)90098-1
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. https://doi.org/10.1038/npre.2010.4282.1
Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends Plant Sci 12:419–426. https://doi.org/10.1016/j.tplants.2007.08.003
Bartoszewski G, Malepszy S, Havey MJ (2004) Mosaic (MSC) cucumbers regenerated from independent cell cultures possess different mitochondrial rearrangements. Curr Genet 45:45–53. https://doi.org/10.1007/s00294-003-0456-6
Bartoszewski G, Havey MJ, Ziółkowska A, Długosz M, Malepszy S (2007) The selection of mosaic (MSC) phenotype after passage of cucumber (Cucumis sativus L.) through cell culture—a method to obtain plant mitochondrial mutants. J Appl Genet 48:1–9. https://doi.org/10.1007/BF03194652
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun 66(10):1148–1152. https://doi.org/10.1107/S1744309110001685
Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot 53(372):1367–1376. https://doi.org/10.1093/jexbot/53.372.1367
Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18:1577–1591. https://doi.org/10.1101/gad.297704
Burke TW, Kadonaga JT (1997) The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Gene Dev 11:3020–3031. https://doi.org/10.1101/gad.11.22.3020
Cassells AC, Curry RF (2001) Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult 64:145–157. https://doi.org/10.1023/A:1010692104861
Chen C, Zhang K, Khurshid M, Li J, He M, Georgiev MI, Zhang X, Zhou M (2019a) MYB transcription repressors regulate plant secondary metabolism. Crit Rev Plant Sci 38(3):159–170. https://doi.org/10.1080/07352689.2019.1632542
Chen S, Zhao H, Luo T, Liu Y, Nie X, Li H (2019b) Characteristics and expression pattern of MYC genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon. Plants 8(8):274. https://doi.org/10.3390/plants8080274
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236. https://doi.org/10.1093/jxb/erh005
Ciereszko I (2018) Regulatory roles of sugars in plant growth and development. Acta Soc Bot Pol 87:3583. https://doi.org/10.5586/asbp.3583
Claaßen C, Gerlach T, Rother D (2019) Stimulus-responsive regulation of enzyme activity for one-step and multi-step syntheses. Adv Synth Catal 361:2387–2401. https://doi.org/10.1002/adsc.201900169
Clausen MV, Hilbers F, Poulsen H (2017) The structure and function of the Na, K-ATPase isoforms in health and disease. Front Physiol 8:371. https://doi.org/10.3389/fphys.2017.00371
Comino C, Moglia A, Repetto A, Tavazza R (2019) Globe Artichoke tissue culture and its biotechnological application, In: Portis E, Acquadro A, Lanteri S (eds) The Globe Artichoke Genome. Compendium of plant genomes. Springer, Cham, pp 41–64. https://doi.org/10.1007/978-3-030-20012-1_3
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
De Assuncao AGL, Martinus GMA (2018) Regulation of zinc deficiency and tolerance in plants. U.S. Patent No. 10,106,810
Dewitte W, Murray JA (2003) The plant cell cycle. Annu Rev Plant Biol 54:235–264. https://doi.org/10.1146/annurev.arplant.54.031902.134836
Ding B, Patterson EL, Holalu SV, Li J, Johnson GA, Stanley LE, Greenlee AB, Peng F, Bradshaw HD Jr, Blinov ML, Blackman BK (2020) Two myb proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr Biol 30(5):802–814. https://doi.org/10.1016/j.cub.2019.12.067
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206. https://doi.org/10.1016/S1360-1385(00)01600-9
Evans DA (1989) Somaclonal variation—genetic basis and breeding applications. Trends Genet 5:46–50. https://doi.org/10.1016/0168-9525(89)90021-8
Fang H, Meng Q, Xu J, Tang H, Tang S, Zhang H, Huang J (2015) Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol Biol 87:441–458. https://doi.org/10.1007/s11103-015-0294-1
Feller A, Machemer K, Braun EL, Grotewold E (2010) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116. https://doi.org/10.1111/j.1365-313X.2010.04459.x
Gu Z, Gu L, Eils R, Schlesner M, Brors B (2014) circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. https://doi.org/10.1093/bioinformatics/btu393
Guo WL, Wu R, Zhang YF, Liu XM, Wang HY, Gong L, Zhang ZH, Liu B (2007) Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth. et Hook. F. Plant Cell Rep 26:1297–1307. https://doi.org/10.1007/s00299-007-0320-0
Guo W, Nazim H, Liang Z, Yang D (2016) Magnesium deficiency in plants: an urgent problem. Crop J 4:83–91. https://doi.org/10.1016/j.cj.2015.11.003
Hao Y-J, Deng X-X (2002) Occurrence of chromosomal variations and plant regeneration from long-term-cultured citrus callus. In Vitro Cell Dev Biol Plant 38:472–476. https://doi.org/10.1079/IVP2002317
Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA (2010) Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137:975–984. https://doi.org/10.1242/dev.047662
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. https://doi.org/10.1016/S1360-1385(01)02223-3
Jevremović S, Subotić A, Miljković D, Trifunović M, Petrić M, Cingel A (2012) Clonal fidelity of chrysanthemum cultivars after long term micropropagation by stem segment culture. VII International symposium on in vitro culture and horticultural breeding. Acta Hortic 961:211–216. https://doi.org/10.17660/ActaHortic.2012.961.25
Karp A (1995) Somaclonal variation as a tool for crop improvement. Euphytica 85:295–302. https://doi.org/10.1007/BF00023959
Khan S, Saeed B, Kauser N (2011) Establishment of genetic fidelity of in-vitro raised banana plantlets. Pak J Bot 43:233–242
Kim JH, Kim J, Jun SE, Park S, Timilsina R, Kwon DS, Kim Y, Park S-J, Hwang JY, Nam HG, Kim G-T, Woo HR (2018) ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol 220:609–623. https://doi.org/10.1111/nph.15291
Krishna H, Alizadeh M, Singh D, Singh U, Chauhan N, Eftekhari M, Sadh RK (2016) Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 6:54. https://doi.org/10.1007/s13205-016-0389-7
Kusnetsov V, Landsberger M, Meurer J, Oelmüller R (1999) The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem 274:36009–36014. https://doi.org/10.1074/jbc.274.50.36009
Larkin PJ, Scowcroft WR (1981) Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:197–214. https://doi.org/10.1007/BF02342540
Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, Ritchie ME (2016) RNA-seq analysis is easy as 1–2-3 with limma, Glimma and edgeR. F1000Research 2016:5. https://doi.org/10.12688/f1000research.9005.3.
Lee-Huang S, Lin J-J, Kung H-F, Huang PL, Lee L, Huang PL (1993) The human erythropoietin-encoding gene contains a CAAT box, TATA boxes and other transcriptional regulatory elements in its 5’ flanking region. Gene 128:227–236. https://doi.org/10.1016/0378-1119(93)90567-M
Lilly JW, Bartoszewski G, Malepszy S, Havey MJ (2001) A major deletion in the cucumber mitochondrial genome sorts with the MSC phenotype. Curr Genet 40:144–151. https://doi.org/10.1007/s002940100238
Liu J, Xia Z, Wang M, Zhang X, Yang T, Wu J (2013) Overexpression of a maize E3 ubiquitin ligase gene enhances drought tolerance through regulating stomatal aperture and antioxidant system in transgenic tobacco. Plant Physiol Biochem 73:114–120. https://doi.org/10.1016/j.plaphy.2013.09.006
Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271:11–21. https://doi.org/10.1007/s00438-003-0954-8
Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950. https://doi.org/10.1105/tpc.022319
Ładyżyński M, Burza W, Malepszy S (2002) Relationship between somaclonal variation and type of culture in cucumber. Euphytica 125:349–356. https://doi.org/10.1023/A:1016017825907
Malepszy S, Burza W, Śmiech M (1996) Characterization of a cucumber (Cucumis sativus L.) somaclonal variant with paternal inheritance. J Appl Genet 37:65–78
Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13:67–73. https://doi.org/10.1016/S0168-9525(96)10049-4
Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends in Biochem Sci 20:506–510. https://doi.org/10.1016/S0968-0004(00)89118-5
Noguero M, Atif RM, Ochatt S, Thompson RD (2013) The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci 209:32–45. https://doi.org/10.1016/j.plantsci.2013.03.016
Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y (2006) The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 27(11):587–593. https://doi.org/10.1016/j.tips.2006.09.001
Osipowski P, Pawełkowicz M, Wojcieszek M, Skarzyńska A, Przybecki Z, Pląder W (2020) A high-quality cucumber genome assembly enhances computational comparative genomics. Mol Genet Genomics 295:177–193. https://doi.org/10.1007/s00438-019-01614-3
Pape SL (2012) EasyqpcR: EasyqpcR for easy analysis of real-time PCR data at IRTOMITINSERM U1082. English. R package. IRTOMIT-INSERM U 1082
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417. https://doi.org/10.1038/nmeth.4197
Pawełkowicz ME, Skarzyńska A, Sroka M, Szwacka M, Pniewski T, Pląder W (2020) Effect of transgenesis on mRNA and miRNA profiles in cucumber fruits expressing Thaumatin II. Genes 11:334. https://doi.org/10.3390/genes11030334
Piagnani C, Iacona C, Intrieri MC, Muleo R (2002) A new somaclone of Prunus Avium shows diverse growth pattern under different spectral quality of radiation. Biol Plantarum 45:11–17. https://doi.org/10.1023/A:1015182608782
Pląder W, Malepszy S, Burza W, Rusinowski Z (1998) The relationship between the regeneration system and genetic variability in the cucumber (Cucumis sativus L.). Euphytica 103:9–15. https://doi.org/10.1023/A:1018359726626
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381. https://doi.org/10.1016/j.tplants.2012.02.004
Ramakers C, Ruijter JM, Deprez RHL, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. https://doi.org/10.1016/S0304-3940(02)01423-4
Schmid KM (2015) Lipid metabolism in plants. In: Ridgway ND, McLeod RS (eds) Biochemistry of lipids, lipoproteins and membranes. Elsevier, Dordrecht, pp 113–147
Sharma B, Joshi D, Yadav PK, Gupta AK, Bhatt TK (2016) Role of ubiquitin-mediated degradation system in plant biology. Front Plant Sci 7:806. https://doi.org/10.3389/fpls.2016.00806
Skarzyńska A, Pawełkowicz M, Pląder W, Przybecki Z (2016) The utility of optical detection system (qPCR) and bioinformatics methods in reference gene expression analysis. In: Photonics applications in astronomy, communications, industry, and high-energy physics experiments, International Society for Optics and Photonics, vol 10031, p 1003130. https://doi.org/10.1117/12.2249147
Skarzyńska A, Pawełkowicz M, Pląder W (2020) Genome-wide discovery of DNA variants in cucumber somaclonal lines. Gene 736:144412. https://doi.org/10.1016/j.gene.2020.144412
Smulders MJM, De Klerk GJ (2011) Epigenetics in plant tissue culture. Plant Growth Regul 63:137–146. https://doi.org/10.1007/s10725-010-9531-4
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45:D362–D368. https://doi.org/10.1093/nar/gkw937
Szwacka M, Morawski M, Burza W, 1996. Agrobacterium tumefaciens-mediated cucumber transformation with thaumatin II cDNA. Journal of Applied Genetics (37A).
Teixeira LK, Reed SI (2013) Ubiquitin ligases and cell cycle control. Annu Rev Biochem 82:387–414. https://doi.org/10.1146/annurev-biochem-060410-105307
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wicker T, Guyot R, Yahiaoui N, Keller B (2003) CACTA transposons in Triticeae. A diverse family of high-copy repetitive elements. Plant Physiol 132:52–63. https://doi.org/10.1104/pp.102.015743
Xu Z-S, Chen M, Li L-C, Ma Y-Z (2008) Functions of the ERF transcription factor family in plants. Botany 86:969–977. https://doi.org/10.1139/B08-041
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685. https://doi.org/10.1111/j.1365-313X.2009.04092.x
Zhang J, Wang S, Song S, Xu F, Pan Y, Wang H (2019) Transcriptomic and proteomic analyses reveal new insight into chlorophyll synthesis and chloroplast structure of maize leaves under zinc deficiency stress. J Proteomics 199:123–134. https://doi.org/10.1016/j.jprot.2019.03.001
Acknowledgements
We thank Prof. Stefan Malepszy (Warsaw University of Life Sciences, Poland) and Prof. Grzegorz Bartoszewski (Warsaw University of Life Sciences, Poland) for providing the seeds. We thank Robert McKenzie, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/ac), for editing a draft of this manuscript.
Funding
This work was supported by grants from the National Science Center (2013/11/B/NZ9/00814 and 2011/01/B/NZ2/01631).
Author information
Authors and Affiliations
Contributions
MP and WP designed and conceived the experiments, MP and WP contributed reagents, materials, and analysis tools. MP, AS, EB, and TM performed the experiments and analyzed the data. MP wrote the manuscript. WP and AS revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Manoj Prasad.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pawełkowicz, M.E., Skarzyńska, A., Mróz, T. et al. Molecular insight into somaclonal variation phenomena from transcriptome profiling of cucumber (Cucumis sativus L.) lines. Plant Cell Tiss Organ Cult 145, 239–259 (2021). https://doi.org/10.1007/s11240-020-02005-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-02005-y