Abstract
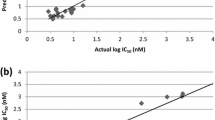
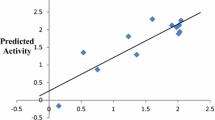
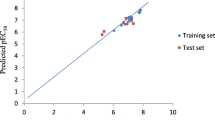
With the aim of researching new antimalarial drugs, a series of quinoline, isoquinoline, and quinazoline derivatives were studied against the Plasmodium falciparum CQ-sensitive and MQ-resistant strain 3D7 protozoan parasite. DFT with B3LYP functional and 6-311G basis set was used to calculate quantum chemical descriptors for QSAR models. The molecular mechanics (MM2) method was used to calculate constitutional, physicochemical, and topological descriptors. By randomly dividing the dataset into training and test sets, we were able to construct reliable models using linear regression (MLR), nonlinear regression (MNLR), and artificial neural networks (ANN). The determination coefficient values indicate the predictive quality of the established models. The robustness and predictive power of the generated models were also confirmed via internal validation, external validation, the Y-randomization test, and the applicability domain. Furthermore, molecular docking studies were conducted to identify the key interactions between the studied molecules and the PfPMT receptor’s active site. The findings of this contribution study indicate that the antimalarial activity of these compounds against Plasmodium falciparum appears to be largely determined by four descriptors, i.e., total connectivity (Tcon), percentage of carbon (C (%)), density (D), and bond length between the two nitrogen atoms (Bond N–N). On the basis of the reliable QSAR model and molecular docking results, several new antimalarial compounds have been designed. The selection of drug candidates was performed according to drug-likeness and ADMET parameters.











Similar content being viewed by others
Availability of data and materials
The data availability is included in the manuscript.
Code availability
Not applicable.
References
Newby G, Bennett A, Larson E et al (2016) The path to eradication: a progress report on the malaria-eliminating countries. Lancet 387:1775–1784. https://doi.org/10.1016/S0140-6736(16)00230-0
Napoli PE, Nioi M (2020) Global Spread of Coronavirus Disease 2019 and Malaria: An Epidemiological Paradox in the Early Stage of A Pandemic. J Clin Med 9:1138. https://doi.org/10.3390/JCM9041138
Mwenesi H, Mbogo C, Casamitjana N et al (2022) Rethinking human resources and capacity building needs for malaria control and elimination in Africa. PLoS Glob Public Heal 2:e0000210. https://doi.org/10.1371/JOURNAL.PGPH.0000210
Hocart SJ, Liu H, Deng H et al (2011) 4-aminoquinolines active against chloroquine-resistant Plasmodium falciparum: basis of antiparasite activity and quantitative structure-activity relationship analyses. Antimicrob Agents Chemother 55:2233–2244. https://doi.org/10.1128/AAC.00675-10
Hadni H, Elhallaoui M (2020) 3D-QSAR, docking and ADMET properties of aurone analogues as antimalarial agents. Heliyon 6:e03580. https://doi.org/10.1016/J.HELIYON.2020.E03580
Hadni H, Mazigh M, Charif E et al (2018) Molecular modeling of antimalarial agents by 3D-QSAR study and molecular docking of two hybrids 4-aminoquinoline-1,3,5-triazine and 4-aminoquinoline-oxalamide derivatives with the receptor protein in its both wild and mutant types. Biochem Res Int 2018. https://doi.org/10.1155/2018/8639173
Hadni H, Elhallaoui M (2020) 2D and 3D-QSAR, molecular docking and ADMET properties in silico studies of azaaurones as antimalarial agents. New J Chem 44:6553–6565. https://doi.org/10.1039/C9NJ05767F
Witola WH, Pessi G, El Bissati K et al (2006) Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus *. J Biol Chem 281:21305–21311. https://doi.org/10.1074/JBC.M603260200
Witola WH, El Bissati K, Pessi G et al (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects *. J Biol Chem 283:27636–27643. https://doi.org/10.1074/JBC.M804360200
Lee SG, Kim Y, Alpert TD et al (2012) Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target. J Biol Chem 287:1426–1434. https://doi.org/10.1074/JBC.M111.315267/ATTACHMENT/F9BD9DDE-084F-4060-81A9-8E07149522FB/MMC2.PDF
Oguike OE, Ugwuishiwu CH, Asogwa CN et al (2022) Systematic review on the application of machine learning to quantitative structure–activity relationship modeling against Plasmodium falciparum. Mol Divers 1:1–16. https://doi.org/10.1007/S11030-022-10380-1/TABLES/1
Aja PM, Agu PC, Ezeh EM et al (2021) (2021) Prospect into therapeutic potentials of Moringa oleifera phytocompounds against cancer upsurge: de novo synthesis of test compounds, molecular docking, and ADMET studies. Bull Natl Res Cent 451(45):1–18. https://doi.org/10.1186/S42269-021-00554-6
Er-rajy M, El Fadili M, Hadni H et al (2022) 2D-QSAR modeling, drug-likeness studies, ADMET prediction, and molecular docking for anti-lung cancer activity of 3-substituted-5-(phenylamino) indolone derivatives. Struct Chem 2022:1–14. https://doi.org/10.1007/S11224-022-01913-3
Kar S, Roy K, Leszczynski J (2022) In silico tools and software to predict ADMET of new drug candidates. 85–115. https://doi.org/10.1007/978-1-0716-1960-5_4
Rasul HO, Aziz BK et al (2022) Correction to: In silico molecular docking and dynamic simulation of eugenol compounds against breast cancer. J Mol Model 2022 284 28:1–1. https://doi.org/10.1007/S00894-022-05068-0
Kpotin GA, Bédé AL, Houngue-Kpota A et al (2019) Relationship between electronic structures and antiplasmodial activities of xanthone derivatives: a 2D-QSAR approach. Struct Chem 30:2301–2310. https://doi.org/10.1007/S11224-019-01333-W/FIGURES/5
Kumari A, Karnatak M, Sen D et al (2022) Synthesis, molecular docking and dynamics study of novel epoxide derivatives of 1,2,4-trioxanes as antimalarial agents. Struct Chem 2022:1–13. https://doi.org/10.1007/S11224-022-01885-4
Ursing J, Johns R, Aydin-Schmidt B et al (2022) Chloroquine-susceptible and -resistant Plasmodium falciparum strains survive high chloroquine concentrations by becoming dormant but are eliminated by prolonged exposure. J Antimicrob Chemother. https://doi.org/10.1093/JAC/DKAC008
Small-Saunders JL, Hagenah LM, Wicht KJ et al (2022) Evidence for the early emergence of piperaquine-resistant Plasmodium falciparum malaria and modeling strategies to mitigate resistance. PLOS Pathog 18:e1010278. https://doi.org/10.1371/JOURNAL.PPAT.1010278
Guillon J, Cohen A, Boudot C et al (2020) Design, synthesis, and antiprotozoal evaluation of new 2,4-bis[(substituted-aminomethyl)phenyl]quinoline, 1,3-bis[(substituted-aminomethyl)phenyl]isoquinoline and 2,4-bis[(substituted-aminomethyl)phenyl]quinazoline derivatives. J Enzyme Inhib Med Chem 35:432–459. https://doi.org/10.1080/14756366.2019.1706502
Chtita S, Ghamali M, Ousaa A et al (2019) QSAR study of anti-Human African Trypanosomiasis activity for 2-phenylimidazopyridines derivatives using DFT and Lipinski’s descriptors. Heliyon 5:e01304. https://doi.org/10.1016/J.HELIYON.2019.E01304
Singh J, Vijay S, Mansuri R et al (2019) Computational and experimental elucidation of Plasmodium falciparum phosphoethanolamine methyltransferase inhibitors: Pivotal drug target. PLoS ONE 14:e0221032. https://doi.org/10.1371/JOURNAL.PONE.0221032
Allinger NL (2002) Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J Am Chem Soc 99:8127–8134. https://doi.org/10.1021/JA00467A001
Strack D (2001) ChemOffice Ultra 2000: CambridgeSoft, c/o CHEM Research GmbH, Frankfurt, DM 3,290. Phytochemistry 57:144. https://doi.org/10.1016/S0031-9422(00)00503-3
Österberg T, Norinder U (2001) Prediction of drug transport processes using simple parameters and PLS statistics The use of ACD/logP and ACD/ChemSketch descriptors. Eur J Pharm Sci 12:327–337. https://doi.org/10.1016/S0928-0987(00)00189-5
Belhassan A, Chtita S, Lakhlifi T, Bouachrine M (2017) QSPR study of the retention/release property of odorant molecules in pectin gels using statistical methods. J Taibah Univ Sci 11:1030–1046. https://doi.org/10.1016/J.JTUSCI.2017.05.004
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA et al, Gaussian09 RA (2009) gaussian. Inc., Wallingford CT, 121, 150–166
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol Biol 1084:193–226. https://doi.org/10.1007/978-1-62703-658-0_11
Lagunin A, Zakharov A, Filimonov D, Poroikov V (2011) QSAR modelling of rat acute toxicity on the basis of PASS prediction. Mol Inform 30:241–250. https://doi.org/10.1002/MINF.201000151
Leonard JT, Roy K (2006) On selection of training and test sets for the development of predictive QSAR models. QSAR Comb Sci 25:235–251. https://doi.org/10.1002/QSAR.200510161
Papa E, Dearden JC, Gramatica P (2007) Linear QSAR regression models for the prediction of bioconcentration factors by physicochemical properties and structural theoretical molecular descriptors. Chemosphere 67:351–358. https://doi.org/10.1016/J.CHEMOSPHERE.2006.09.079
Salt DW, Yildiz N, Livingstone DJ, Tinsley CJ (1992) The use of artificial neural networks in QSAR. Pestic Sci 36:161–170. https://doi.org/10.1002/PS.2780360212
Vidal NP, Manful CF, Pham TH et al (2020) The use of XLSTAT in conducting principal component analysis (PCA) when evaluating the relationships between sensory and quality attributes in grilled foods. MethodsX 7:100835. https://doi.org/10.1016/J.MEX.2020.100835
Wang WW, Ni RQ, Yu FY et al (2018) Optimization of GERD therapeutic regimen based on ANN and realization of MATLAB. Digit Chinese Med 1:47–55. https://doi.org/10.1016/S2589-3777(19)30007-2
Kiralj R, Ferreira MMC (2009) Basic validation procedures for regression models in QSAR and QSPR studies: theory and application. J Braz Chem Soc 20:770–787. https://doi.org/10.1590/S0103-50532009000400021
Gramatica P, Sangion A (2016) A historical excursus on the statistical validation parameters for QSAR Models: a clarification concerning metrics and terminology. J Chem Inf Model 56:1127–1131. https://doi.org/10.1021/ACS.JCIM.6B00088
Javidfar M, Ahmadi S (2020) QSAR modelling of larvicidal phytocompounds against Aedes aegypti using index of ideality of correlation. 31:717–739. https://doi.org/10.1080/1062936X.2020.1806922
Rácz A, Bajusz D, Héberger K (2018) Modelling methods and cross-validation variants in QSAR: a multi-level analysis. 29:661–674. https://doi.org/10.1080/1062936X.2018.1505778
Kůrková V (1992) Kolmogorov’s theorem and multilayer neural networks. Neural Netw 5:501–506. https://doi.org/10.1016/0893-6080(92)90012-8
Andrea TA, Kalayeh H (1991) Applications of neural networks in quantitative structure-activity relationships of dihydrofolate reductase inhibitors. J Med Chem 34:2824–2836. https://doi.org/10.1021/JM00113A022
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20:269–276. https://doi.org/10.1016/S1093-3263(01)00123-1
Rücker C, Rücker G, Meringer M (2007) y-Randomization and its variants in QSPR/QSAR. J Chem Inf Model 47:2345–2357. https://doi.org/10.1021/CI700157B
Roy K, Mitra I (2011) On various metrics used for validation of predictive QSAR models with applications in virtual screening and focused library design. Comb Chem High Throughput Screen 14:450–474. https://doi.org/10.2174/138620711795767893
Netzeva TI, Worth AP, Aldenberg T et al (2005) Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships. The report and recommendations of ECVAM Workshop 52. Altern Lab Anim 33:155–173. https://doi.org/10.1177/026119290503300209
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR Comb Sci 26:694–701. https://doi.org/10.1002/QSAR.200610151
Eriksson L, Jaworska J, Worth AP et al (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ Health Perspect 111:1361. https://doi.org/10.1289/EHP.5758
Frey A, Di Canzio J, Zurakowski D (1998) A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods 221:35–41. https://doi.org/10.1016/S0022-1759(98)00170-7
Hansch C, Leo A, Mekapati SB, Kurup A (2004) QSAR and ADME. Bioorg Med Chem 12:3391–3400. https://doi.org/10.1016/J.BMC.2003.11.037
Jin Z, Wang Y, Yu XF et al (2020) Structure-based virtual screening of influenza virus RNA polymerase inhibitors from natural compounds: Molecular dynamics simulation and MM-GBSA calculation. Comput Biol Chem 85. https://doi.org/10.1016/J.COMPBIOLCHEM.2020.107241
Chtita S, Aouidate A, Belhassan A et al (2020) QSAR study of N-substituted oseltamivir derivatives as potent avian influenza virus H5N1 inhibitors using quantum chemical descriptors and statistical methods. New J Chem 44:1747–1760. https://doi.org/10.1039/C9NJ04909F
Daina A, Michielin O (2017) Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Reports 71(7):1–13. https://doi.org/10.1038/srep42717
Pires DEV, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/ACS.JMEDCHEM.5B00104/SUPPL_FILE/JM5B00104_SI_001.PDF
Chtita S, Belhassan A, Aouidate A et al (2021) Discovery of potent SARS-CoV-2 inhibitors from approved antiviral drugs via docking and virtual screening. Comb Chem High Throughput Screen 24:441–454. https://doi.org/10.2174/1386207323999200730205447
Wang Q, He J, Wu D et al (2015) Interaction of α-cyperone with human serum albumin: Determination of the binding site by using Discovery Studio and via spectroscopic methods. J Lumin 164:81–85. https://doi.org/10.1016/J.JLUMIN.2015.03.025
Sriramulu DK, Wu S, Lee SG (2020) Effect of ligand torsion number on the AutoDock mediated prediction of protein-ligand binding affinity. J Ind Eng Chem 83:359–365. https://doi.org/10.1016/J.JIEC.2019.12.009
Verdonk ML, Cole JC, Hartshorn MJ et al (2003) Improved protein-ligand docking using GOLD. Proteins Struct Funct Genet 52:609–623. https://doi.org/10.1002/PROT.10465
Westermaier Y, Barril X, Scapozza L (2015) Virtual screening: an in silico tool for interlacing the chemical universe with the proteome. Methods 71:44–57. https://doi.org/10.1016/J.YMETH.2014.08.001
Roy K, Kar S, Ambure P (2015) On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab Syst 145:22–29. https://doi.org/10.1016/J.CHEMOLAB.2015.04.013
OECD principles for the validation, for regulatory purposes, of (quantitative) structure-activity relationship models
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inform 29:476–488. https://doi.org/10.1002/MINF.201000061
Belhassan A, Chtita S, Lakhlifi T, Bouachrine M (2017) QSPR study of the retention/release property of odorant molecules in water using statistical methods. Orbital 9:234–247. https://doi.org/10.17807/ORBITAL.V9I4.978
Garg R, Smith CJ (2014) Predicting the bioconcentration factor of highly hydrophobic organic chemicals. Food Chem Toxicol 69:252–259. https://doi.org/10.1016/J.FCT.2014.03.035
Irwin JJ, Tang KG, Young J et al (2020) ZINC20 - a free ultralarge-scale chemical database for ligand discovery. J Chem Inf Model 60:6065–6073. https://doi.org/10.1021/ACS.JCIM.0C00675/SUPPL_FILE/CI0C00675_SI_001.PDF
Kalantzi L, Goumas K, Kalioras V et al (2006) Characterization of the human upper gastrointestinal contents under conditions simulating bioavailability/bioequivalence studies. Pharm Res 23:165–176. https://doi.org/10.1007/S11095-005-8476-1
Speciale A, Muscarà C, Molonia MS et al (2021) Silibinin as potential tool against SARS-Cov-2: in silico spike receptor-binding domain and main protease molecular docking analysis, and in vitro endothelial protective effects. Phytother Res 35:4616–4625. https://doi.org/10.1002/PTR.7107
Han Y, Zhang J, Hu CQ et al (2019) In silico ADME and toxicity prediction of ceftazidime and its impurities. Front Pharmacol 10:434. https://doi.org/10.3389/FPHAR.2019.00434/BIBTEX
Domínguez-Villa FX, Durán-Iturbide NA, Ávila-Zárraga JG (2021) Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl)indol-4-ones: potential inhibitors of SARS CoV-2 main protease. Bioorg Chem 106:104497. https://doi.org/10.1016/J.BIOORG.2020.104497
Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. https://doi.org/10.1016/J.PHARMTHERA.2012.12.007
Ferraz ERA, Umbuzeiro GA, de-Almeida G, et al (2011) Differential toxicity of Disperse Red 1 and Disperse Red 13 in the Ames test, HepG2 cytotoxicity assay, and Daphnia acute toxicity test. Environ Toxicol 26:489–497. https://doi.org/10.1002/TOX.20576
Pogliani L (2000) From molecular connectivity indices to semiempirical connectivity terms: recent trends in graph theoretical descriptors. Chem Rev 100:3827–3858. https://doi.org/10.1021/CR0004456
Acknowledgements
We are grateful to the “Association Marocaine des Chimistes Théoriciens” (AMCT) for its pertinent help concerning the programs and software.
Author information
Authors and Affiliations
Contributions
Said El Rhabori, Abdellah El Aissouq, Samir Chtita, and Fouad Khalil: these authors contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
El Rhabori, S., El Aissouq, A., Chtita, S. et al. QSAR, molecular docking and ADMET studies of quinoline, isoquinoline and quinazoline derivatives against Plasmodium falciparum malaria. Struct Chem 34, 585–603 (2023). https://doi.org/10.1007/s11224-022-01988-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-022-01988-y