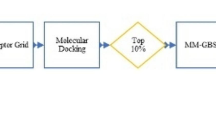
Abstract
HIV protease inhibitors are one of the most important agents for the treatment of HIV infection. In this work, molecular modeling studies combining 3D-QSAR, molecular docking, MESP, HOMO, and LUMO energy calculations were performed on propiophenone derivatives to explore structure activity relationships and structural requirements for the inhibitory activity. The aim of this study was to create a field point–based 3D-QSAR (3D-Quantitative structure-activity relationship) model by using chalcone structures with anti-HIV-1 protease activity from our previous study and to design new potentially more potent and safer inhibitors. The developed model showed acceptable predictive and descriptive capability as represented by standard statistical parameters R2 (0.94) and Q2 (0.59). High correlation between experimental and predicted activities of training set is noticed. All compounds fit into the defined applicability domain. The derived pharmacophoric features were further supported by MESP and Mulliken charge analysis using density functional theory. Statistically significant variables from 3D-QSAR were used to define key structural characteristics which enhance anti-HIV-1 protease activity. This information has been used to design new structures with anti-HIV-1 protease activity. Docking studies were conducted to understand the interactions in predicted compounds. All the compounds were subjected to in silico ADMET profiling in order to select the best potential drug candidates.









Similar content being viewed by others
Availability of data and material
The data that supports the findings of this study are available within the article.
References
UNAIDS [homepage on the Internet] UNAIDS (2020); c2020 [cited 2020 May 10]. Available from: https://www.unaids.org/en
Sharp PM, Hahn BH (2011) Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:1–22. https://doi.org/10.1101/cshperspect.a006841
Louten J (2016) Human immunodeficiency virus. Essential Human Virology; Elsevier Inc. 193–211
Das Neves J (2019) Novel approaches for the delivery of anti-hiv drugs—what is new? Pharmaceutics 11:554. https://doi.org/10.3390/pharmaceutics11110554
Sahu NK, Balbhadra SS, Choudhary J, Kohli DV (2019) Exploring pharmacological significance of chalcone scaffold: a review. Curr Med Chem 19:209–225. https://doi.org/10.2174/092986712803414132
Nowakowska Z (2007) A review of anti-infective and anti-inflammatory chalcones. Eur J Med Chem 42:125–37. https://doi.org/10.1016/j.ejmech.2006.09.019
Babasaheb PB, Shrikant SG, Ragini GB, Jalinder VT, Chandrahas NK (2010) Synthesis and biological evaluation of simple methoxylated chalcones asanticancer, anti-inflammatory and antioxidant agents. Bioorg Med Chem 18:1364–1370. https://doi.org/10.1016/j.bmc.2009.11.066
Ansari FL, Nazir S, Noureen H, Mirza B (2005) Combinatorial synthesis and antibacterial evaluation of an indexed chalcone library. Chem Biodivers 2:1656–64. https://doi.org/10.1002/cbdv.200590135
Lahtchev KL, Batovska DI, Parushev SP, Ubiyvovk VM, Sibirny AA (2008) Antifungal activitymof chalcones: a mechanistic study using various yeast strains. Eur J Med Chem 43:2220–8. https://doi.org/10.1016/j.ejmech.2007.12.027
Dimmock JR, Kandepu NM, Hetherington M, Quail WJ, Pugazhenthi U, Sudom AM et al (1998) Cytotoxic activities of Mannich bases of chalcones and related compounds. J Med Chem 41:1014–26. https://doi.org/10.1021/jm970432t
Lin YM, Zhou Y, Flavin MT, Zhou LM, Nie W, Chen FC (2002) Chalcones and flavonoids as anti-tuberculosis agents. Bioorg Med Chem 10:2795–802. https://doi.org/10.1016/s0968-0896(02)00094-9
Mateeva N, Eyunni SVK, Redda KK, Ononuju U, Hansberry TD, Aikens C, Nag A (2017) Functional evaluation of synthetic flavonoids and chalcones for potential antiviral and anticancer properties. Bioorg Med Chem Lett 27:2350–2356. https://doi.org/10.1016/j.bmcl.2017.04.034
Cole AL, Hossain S, Cole AM, Phanstiel O IV (2016) Synthesis and bioevaluation of substituted chalcones, coumaranones and other flavonoids as anti-HIV agents. Bioorg Med Chem 24:2768–76. https://doi.org/10.1016/j.bmc.2016.04.045
Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in drug design - a review. Curr Top Med Chem 10:95–115. https://doi.org/10.2174/156802610790232260
Chen Z, Li P, Hu D et al (2019) Synthesis, antiviral activity, and 3D-QSAR study of novel chalcone derivatives containing malonate and pyridine moieties. Arab J Chem 12:2685–2696. https://doi.org/10.1016/j.arabjc.2015.05.003
Wan Z, Hu D, Li P, Xie D, Gan X (2015) Synthesis, antiviral bioactivity of novel 4-thioquinazoline derivatives containing chalcone moiety. Molecules 20:11861–11874. https://doi.org/10.3390/molecules200711861
Sharma H, Patil S, Sanchez TW, Neamati N, Schinazi RF, Buolamwini JK (2011) Synthesis, biological evaluation and 3D-QSAR studies of 3-keto salicylic acid chalcones and related amides as novel HIV-1 integrase inhibitors. Bioorg Med Chem 19:2030–2045. https://doi.org/10.1016/j.bmc.2011.01.047
Turkovic N, Ivkovic B, Kotur-Stevuljevic J, Tasic M, Marković B, Vujic Z (2020) Molecular docking, synthesis and anti-HIV-1 protease activity of novel chalcones. Current Pharmaceutical Design 26:802–814. https://doi.org/10.2174/1381612826666200203125557
Tomasi J, Mennucci B, Cammi R (2005) Quantum mechanical continuum solvatation models. Chem Rev 105:2999–3093. https://doi.org/10.1021/cr9904009
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery Jr JA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant J C, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09 (Revision D.01), Gaussian, Inc., Wallingford CT
Pentacle V (2009) 1.0.7 Molecular Discovery Ltd., Perugia, Italy. http://www.moldiscovery.com/software/pentacle/
Durán Á, Martínez GC, Pastor M (2008) Development and validation of AMANDA, a new algorithm for selecting highly relevant regions in molecular interaction fields. J Chem Inf Model 48:1813–1823. https://doi.org/10.1021/ci800037t
Durán Á, Zamora I, Pastor M (2009) Suitability of GRIND-based principal properties for the description of molecular similarity and ligand-based virtual screening. J Chem Inf Model 49:2129–2138. https://doi.org/10.1021/ci900228x
Ojha PK, Roy K (2011) Comparative QSARs for antimalarial endochins: Importance of descriptor-thinning and noise reduction prior to feature selection. Chemom Intell Lab Syst 109:146–161. https://doi.org/10.1016/j.chemolab.2011.08.007
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inform 29:476–488. https://doi.org/10.1002/minf.201000061
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring rm2 metrics for validation of QSPR models. Chemom Intell Lab Syst 107:194–205. https://doi.org/10.1016/j.chemolab.2011.03.011
Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52:396–408. https://doi.org/10.1021/ci200520g
OECD (2014) Guidance document on the validation of (quantitative) structure-activity relationship [(Q)SAR] models, OECD Series on Testing and Assessment, No. 69, OECD Publishing, Paris. https://doi.org/10.1787/9789264085442-en
Roy K, Kar S, Das RN (2015) A primer on QSAR/QSPR modeling. SpringerBriefs Mol Sci
SPSS Inc. Released (2009) PASW Statistics for Windows, version 18.0. Chicago: SPSS Inc
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR & Combinatorial Science 26:694–701. https://doi.org/10.1002/qsar.200610151
Ghosh J, Lawless MS, Waldman M, Gombar V, Fraczkiewicz R (2016) Modeling ADMET. Methods Mol Biol (Clifton, N.J.) 1425:63–83. https://doi.org/10.1007/978-1-4939-3609-0_4
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–61. https://doi.org/10.1002/jcc.21334
BOVIA, Dassault Systèmes, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes (2016). https://discover.3ds.com/discovery-studio-visualizer-download
Acknowledgements
We thank the COST Action CA17120 Chemobrionics (CBrio) of the European Community for support.
Funding
This research was funded by the Ministry of Education, Science and Technological Development of the Republic of Serbia grant number 451-03-9/2021-14/200026. S.G. received support from the Serbian Ministry of Education and Science (Grant No. 451-03-9/2021-14/200026).
Author information
Authors and Affiliations
Contributions
M.J., K.N., and N.T. contributed toward the 3DQSAR and ADMET analysis, as well as the interpretation of the relevant results. M.J. contributed also to the docking studies. S.G. performed the DFT calculations, contributed to the docking studies, and interpreted the results. S.G., B.I., and Z.V. coordinated the research and wrote the final manuscript based on the research reports. All the authors exchanged opinions concerning the progress of the project and commented on the preparation of the manuscript at all stages. All the authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jovanović, M., Turković, N., Ivković, B. et al. 3D-QSAR, molecular docking and in silico ADMET studies of propiophenone derivatives with anti-HIV-1 protease activity. Struct Chem 32, 2341–2353 (2021). https://doi.org/10.1007/s11224-021-01810-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-021-01810-1