Abstract
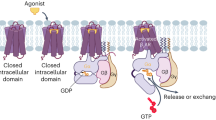
G-protein–coupled receptors are integral membrane proteins involved in signal transduction pathways, making them an appealing drug targets for a wide spectrum of diseases. The previous literature reports provide an evidence that catecholamine regulates metastasis by actuating the β2-adrenergic receptor (β-2AR). Molecular dynamics simulations were carried out for 1000 ns to understand the effect of the catecholamine on the human β-2AR. On comparing the apoprotein structure of β-2AR with that of catecholamine interacted β-2AR protein, a large change in structural assembly is observed in the helical regions which confirm the activation of β-2AR protein. The visualization of internal natural pathway of β-2AR protein structure gives us detailed information about deviation in TM helixes. The compactness of protein structure shows β-2AR protein interacting with epinephrine is much stable than β-2AR protein interacting with norepinephrine structure. The Gibbs free energy shows norepinephrine as a partial agonist whereas epinephrine as full agonist for β-2AR protein.














Similar content being viewed by others
Abbreviations
- GPCRs:
-
G-protein coupled receptors
- SNS:
-
Sympathetic nervous system
- MD:
-
Molecular dynamics
- β-2AR:
-
β2-adrenergic receptor
- TM:
-
Transmembrane
- CNS:
-
Central nervous system
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- Rg:
-
Radius of gyration
- SASA:
-
Solvent accessible surface area
- FEL:
-
Free energy landscape
References
McLaughlin JT, Fu J, Rosenberg RL (2007) Agonist-driven conformational changes in the inner beta-sheet of alpha7 nicotinic receptors. Mol Pharmacol 71:1312–1318. https://doi.org/10.1124/mol.106.033092.loop
Kobilka BK (2007) G protein coupled receptor structure and activation. Biochim Biophys Acta - Biomembr 1768:794–807. https://doi.org/10.1016/j.bbamem.2006.10.021
Isin B, Estiu G, Wiest O, Oltvai ZN (2012) Identifying ligand binding conformations of the ??2-adrenergic receptor by using its agonists as computational probes. PLoS One 7. https://doi.org/10.1371/journal.pone.0050186
Lefkowitz RJ, Sun J, Shukla AK (2008) A crystal clear view of the beta2-adrenergic receptor. Nat Biotechnol 26:189–191. https://doi.org/10.1038/nbt0208-189
Bang I, Choi H-J (2015) Structural features of β-2 adrenergic receptor: crystal structures and beyond overall structure of β-2 adrenergic receptor molecules and cells. Mol Cells 38:105–111. https://doi.org/10.14348/molcells.2015.2301
Bruno A, Costantino G (2012) Molecular dynamics simulations of g protein-coupled receptors. Mol Inform 31:222–230. https://doi.org/10.1002/minf.201100138
Goddard WA, Abrol R (2007) 3-Dimensional structures of G protein-coupled receptors and binding sites of agonists and antagonists. J Nutr 137:1528S–1538S. https://doi.org/10.1093/jn/137.6.1528s
Bang I, Choi H-J (2015) Structural features of β2 adrenergic receptor: crystal structures and beyond. Mol Cells 38:105–111. https://doi.org/10.14348/molcells.2015.2301
Balaraman GS, Bhattacharya S, Vaidehi N (2010) Structural insights into conformational stability of wild-type and mutant β1-adrenergic receptor. Biophys J 99:568–577. https://doi.org/10.1016/j.bpj.2010.04.075
Hanlon CD, Andrew DJ (2015) Outside-in signaling - a brief review of GPCR signaling with a focus on the Drosophila GPCR family. J Cell Sci 128:3533–3542. https://doi.org/10.1242/jcs.175158
Kuzumaki N, Suzuki A, Narita M, et al (2012) Multiple analyses of G-protein coupled receptor (GPCR) expression in the development of gefitinib-resistance in transforming non-small-cell lung cancer. PLoS One 7:. https://doi.org/10.1371/journal.pone.0044368
Due M, Khanna M, Wang R et al (2019) Catalytic triad heterogeneity in S51 peptidase family : structural basis for functional variability. Nat Publ Gr 6:56562–56570. https://doi.org/10.1002/prot.25693
Hwa Jin Lee, Brian Wall and SC (2005) G-protein-coupled receptors and melanoma. Pigment Cell Melanoma Res 23:1–7. https://doi.org/10.1038/jid.2014.371
Bhola NE, Grandis JR (2008) Crosstalk between G-protein-coupled receptors and epidermal growth factor receptor in cancer. Front Biosci a J virtual Libr 13:1857–1865
Bauknecht P, Jékely G (2017) Ancient coexistence of norepinephrine, tyramine, and octopamine signaling in bilaterians. BMC Biol 15:6. https://doi.org/10.1186/s12915-016-0341-7
Lodish H, Berk A, Zipursky SL et al (2000) Molecular cell biology4th edn. Natl Cent Biotechnol InformationÕs Bookshelf
Kim T-H, Rowat AC, Sloan EK (2016) Neural regulation of cancer: from mechanobiology to inflammation. Clin Transl Immunol 5:e78. https://doi.org/10.1038/cti.2016.18
Ozcan O, Uyar A, Doruker P, Akten E (2013) Effect of intracellular loop 3 on intrinsic dynamics of human β2-adrenergic receptor. BMC Struct Biol 13:29. https://doi.org/10.1186/1472-6807-13-29
Bourlat SJ, Juliusdottir T, Lowe CJ et al (2006) Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444:85–88. https://doi.org/10.1038/nature05241
S RL and D (2015) Neural regulation of hematopoiesis, inflammation and cancer. Neuron 176:139–148. https://doi.org/10.1016/j.physbeh.2017.03.040
Wang Y, De Arcangelis V, Gao X et al (2008) Norepinephrine- and epinephrine-induced distinct β2- adrenoceptor signaling is dictated by GRK2 phosphorylation in cardiomyocytes. J Biol Chem 283:1799–1807. https://doi.org/10.1074/jbc.M705747200
Blais V, Bounif N, Dubé F (2010) Characterization of a novel octopamine receptor expressed in the surf clam Spisula solidissima. Gen Comp Endocrinol 167:215–227. https://doi.org/10.1016/j.ygcen.2010.03.008
Makita N, Kabasawa Y, Otani Y et al (2013) Attenuated desensitization of β-adrenergic receptor by water-soluble N-nitrosamines that induce S-nitrosylation without NO release. Circ Res 112:327–334. https://doi.org/10.1161/CIRCRESAHA.112.277665
Bornert O, Møller TC, Boeuf J, et al (2013) Identification of a novel protein-protein interaction motif mediating interaction of GPCR-associated sorting proteins with G protein-coupled receptors. PLoS One 8:. https://doi.org/10.1371/journal.pone.0056336
de Lucia C, Eguchi A, Koch WJ (2018) New insights in cardiac β-Adrenergic signaling during heart failure and aging. Front Pharmacol 9:1–14. https://doi.org/10.3389/fphar.2018.00904
Whalen EJ, Foster MW, Matsumoto A et al (2007) Regulation of β-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell 129:511–522. https://doi.org/10.1016/j.cell.2007.02.046
Schledorn M, Meier BH, Böckmann A et al (2018) Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Biochemistry 7:269–277. https://doi.org/10.3389/fpubh.2018.00095
Vanni S, Neri M, Tavernelli I, Rothlisberger U (2009) Observation of “ionic lock” formation in molecular dynamics simulations of wild-type β1 and β2 adrenergic receptors. Biochemistry 48:4789–4797. https://doi.org/10.1021/bi900299f
Brody T, Cravchik A (2000) Drosophila melanogaster G protein-coupled receptors. J Cell Biol 150:F83–F88. https://doi.org/10.1083/jcb.150.2.F83
Chitrala KN, Yeguvapalli S (2014) Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 gene. PLoS One 9:. https://doi.org/10.1371/journal.pone.0104242
Kmiecik S, Jamroz M, Kolinski M (2014) Structure prediction of the second extracellular loop in G-protein-coupled receptors. Biophys J 106:2408–2416. https://doi.org/10.1016/j.bpj.2014.04.022
Rasmussen SGF, Choi H-J, Fung JJ et al (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469:175–180. https://doi.org/10.1038/nature09648
Wang T, Duan Y (2009) Ligand entry and exit pathways in the β2-adrenergic receptor. J Mol Biol 392:1102–1115. https://doi.org/10.1016/j.jmb.2009.07.093
Trzaskowski B, Latek D, Yuan S et al (2012) Action of molecular switches in GPCRs - theoretical and experimental studies. Curr Med Chem 19:1090–1109. https://doi.org/10.2174/092986712799320556
Grossfield A (2011) Recent progress in the study of G protein-coupled receptors with molecular dynamics computer simulations. Biochim Biophys Acta - Biomembr 1808:1868–1878. https://doi.org/10.1016/j.bbamem.2011.03.010
Shahane G, Parsania C, Sengupta D, Joshi M (2014) Molecular insights into the dynamics of pharmacogenetically important N-terminal variants of the human β2-adrenergic receptor. PLoS Comput Biol 10. https://doi.org/10.1371/journal.pcbi.1004006
Gouldson PR, Snell CR, Reynolds CA (1997) A new approach to docking in the β2-adrenergic receptor that exploits the domain structure of G-protein-coupled receptors. J Med Chem 40:3871–3886. https://doi.org/10.1021/jm960647n
Strader CD, Fong TM, Tota MR et al (1994) Structure and function of G protein-coupled receptors. Annu Rev Biochem 63:101–132
Maïga A, Dupont M, Blanchet G et al (2014) Molecular exploration of the α1A-adrenoceptor orthosteric site: binding site definition for epinephrine, HEAT and prazosin. FEBS Lett 588:4613–4619. https://doi.org/10.1016/j.febslet.2014.10.033
Spijker P, Vaidehi N, Freddolino PL et al (2006) Dynamic behavior of fully solvated ?2-adrenergic receptor, embedded in the membrane with bound agonist or antagonist. Proc Natl Acad Sci U S A 103:4882–4887. https://doi.org/10.1073/pnas.0511329103
Periole X (2017) Interplay of G protein-coupled receptors with the membrane: Insights from supra-atomic coarse grain molecular dynamics simulations. Chem Rev 117:156–185. https://doi.org/10.1021/acs.chemrev.6b00344
Huber T, Menon S, Sakmar TP (2008) Structural basis for ligand binding and specificity in adrenergic receptors: implications for GPCR-targeted drug discovery. Biochemistry 47:11013–11023. https://doi.org/10.1021/bi800891r
Wolf S, Böckmann M, Höweler U et al (2008) Simulations of a G protein-coupled receptor homology model predict dynamic features and a ligand binding site. FEBS Lett 582:3335–3342. https://doi.org/10.1016/j.febslet.2008.08.022
Cryer PE, Haymond MW, Santiago JV, Shah SD (1976) Norepinephrine and epinephrine release and adrenergic mediation of smoking-associated hemodynamic and metabolic events. N Engl J Med 295:573–577. https://doi.org/10.1056/nejm197609092951101
Roth KA, Mefford IM, Barchas JD (1982) Epinephrine, norepinephrine, dopamine and serotonin: differential effects of acute and chronic stress on regional brain amines. Brain Res 239:417–424
Cherezov V, Rosenbaum DM, Hanson MA, et al (2007) High-resolution crystal structure of an engineered human b 2 -adrenergic. Science (80- ) 318:1258–1265
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Sterling T, Irwin JJ (2015) ZINC 15 - Ligand discovery for everyone. J Chem Inf Model 55:2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Jing Huang and Alexander D. Mackerell Jr (2015) NIH Public Access. 34:213–223. https://doi.org/10.1007/978-1-62703-673-3
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/jcc.21367
Mayne CG, Saam J, Schulten K et al (2013) Rapid parameterization of small molecules using the force field toolkit. J Comput Chem 34:2757–2770
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Trott O, Olson A (2010) NIH public access. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334.AutoDock
Furse KE, Lybrand TP (2003) Three-dimensional models for β-adrenergic receptor complexes with agonists and antagonists. J Med Chem 46:4450–4462. https://doi.org/10.1021/jm0301437
Abraham MJ, Murtola T, Schulz R et al (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Hess B, Kutzner C, Van Der Spoel D, Lindahl E (2008) GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. https://doi.org/10.1021/ct700301q
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29:1859–1865
Boonstra S, Onck PR, Van Der Giessen E (2016) CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state. J Phys Chem B 120:3692–3698. https://doi.org/10.1021/acs.jpcb.6b01316
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A 105:9954–9960. https://doi.org/10.1021/jp003020w
Berendsen HJC, Postma JPM, van Gunsteren WF et al (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N·log( N ) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Fadrná E, Hladecková K, Koca J (2005) Long-range electrostatic interactions in molecular dynamics: an endothelin-1 case study. J Biomol Struct Dyn 23:151–162. https://doi.org/10.1080/07391102.2005.10531229
Van Gunsteren WF, Berendsen HJC (1988) A leap-frog algorithm for stochastic dynamics. Mol Simul 1:173–185
Morris GM, Goodsell DS, Halliday RS, et al (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662. 10.1002/(SICI)1096-987X(19981115)19:14 < 1639::AID-JCC10 > 3.0.CO;2-B
Huang K, García AE (2014) Effects of truncating van der Waals interactions in lipid bilayer simulations. J Chem Phys 141:1–9. https://doi.org/10.1063/1.4893965
Bjelkmar P, Larsson P, Cuendet MA et al (2010) Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput 6:459–466. https://doi.org/10.1021/ct900549r
Carugo O, Pongor S (2008) A normalized root-mean-spuare distance for comparing protein three-dimensional structures. Protein Sci 10:1470–1473. https://doi.org/10.1110/ps.690101
Kufareva I, Abagyan R (2012) Homology modeling. 857:231–257. https://doi.org/10.1007/978-1-61779-588-6
Schneider M, Wolf S, Schlitter J, Gerwert K (2011) The structure of active opsin as a basis for identification of GPCR agonists by dynamic homology modelling and virtual screening assays. FEBS Lett 585:3587–3592. https://doi.org/10.1016/j.febslet.2011.10.027
Zhang R, Ming Y, Liu Y (2015) Long time molecular dynamic simulation on the agonist binding and activation of the β 2-adrenergic receptor. Mol. Simul. 41:564–571
Goetz A, Lanig H, Gmeiner P, Clark T (2011) Molecular dynamics simulations of the effect of the G-protein and diffusible ligands on the β2-adrenergic receptor. J Mol Biol 414:611–623. https://doi.org/10.1016/j.jmb.2011.10.015
Katritch V, Abagyan R (2011) GPCR agonist binding revealed by modeling and crystallography. Trends Pharmacol Sci 32:637–643. https://doi.org/10.1016/j.tips.2011.08.001
Bornot A, Etchebest C, De Brevern AG (2011) Predicting protein flexibility through the prediction of local structures. Proteins Struct Funct Bioinforma 79:839–852. https://doi.org/10.1002/prot.22922
Ozcan O, Uyar A, Doruker P, Akten ED (2013) Effect of intracellular loop 3 on intrinsic dynamics of human β 2 -adrenergic receptor effect of intracellular loop 3 on intrinsic dynamics of human β 2 -adrenergic receptor. BMC Struct Biol
Chung KY, Rasmussen SGF, Liu T et al (2011) Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature 477:611–615. https://doi.org/10.1038/nature10488
Keerthana SP, Kolandaivel P (2015) Study of mutation and misfolding of Cu-Zn SOD1 protein. J Biomol Struct Dyn 33:167–183. https://doi.org/10.1080/07391102.2013.865104
Arnold WD, Oldfield E (2000) The chemical nature of hydrogen bonding in proteins via NMR: J-couplings, chemical shifts, and AIM theory. J Am Chem Soc 122:12835–12841. https://doi.org/10.1021/ja0025705
Pace CN, Fu H, Fryar KL et al (2014) Contribution of hydrogen bonds to protein stability. Protein Sci 23:652–661. https://doi.org/10.1002/pro.2449
Nagasundaram N, Zhu H, Liu J et al (2015) Analysing the effect of mutation on protein function and discovering potential inhibitors of CDK4: molecular modelling and dynamics studies. PLoS One 10:1–21. https://doi.org/10.1371/journal.pone.0133969
Durham E, Dorr B, Woetzel N et al (2009) Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J Mol Model 15:1093–1108. https://doi.org/10.1007/s00894-009-0454-9
Pellegrini-Calace M, Maiwald T, Thornton JM (2009) PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLoS Comput Biol 5. https://doi.org/10.1371/journal.pcbi.1000440
Kohlhoff KJ, Shukla D, Lawrenz M et al (2014) Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chem 6:15–21. https://doi.org/10.1038/nchem.1821
Tomobe K, Yamamoto E, Kholmurodov K, Yasuoka K (2017) Water permeation through the internal water pathway in activated GPCR rhodopsin. PLoS One 12:e0176876
Kabsch W, Sander C (1983) Dictionary of protein secondary structure - pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637. https://doi.org/10.1002/bip.360221211
Revision D, Frisch MJ, Trucks GW et al (2015) Gaussian 09. Revision. 1–20
Boys S, Bernardi F (1970) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys 19:553–566. https://doi.org/10.1080/00268977000101561
Reiner S, Ambrosio M, Hoffmann C, Lohse MJ (2010) Differential signaling of the endogenous agonists at the β2-adrenergic receptor. J Biol Chem 285:36188–36198. https://doi.org/10.1074/jbc.M110.175604
Funding
I Rahul Suresh, express my sincere thanks to DST-PURSE for the financial assistant provided during this course of work.
Author information
Authors and Affiliations
Contributions
All co-authors of the submitted manuscript titled “Molecular dynamics simulation involved in expounding the activation of adrenoceptors by sympathetic nervous system signaling” are aware and have approved the submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 204 kb)
Rights and permissions
About this article
Cite this article
Suresh, R., Subramaniam, V. Molecular dynamics simulation involved in expounding the activation of adrenoceptors by sympathetic nervous system signaling. Struct Chem 31, 1869–1885 (2020). https://doi.org/10.1007/s11224-020-01553-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01553-5