Abstract
The Quantitative Property-Retention Relation (QPRR) approach was applied to analyze the correlations between the retention parameters of ampholytic, biologically active substances and their physicochemical (predicted/spectral) characteristics. The retention parameters were obtained for polar and semipolar HPLC columns at various compositions of mobile phases and pH conditions. These values are a unique collection of chromatographic parameters that are a measure of lipophilicity and, consequently, can be very helpful in assessing pharmacological potency of the compounds investigated. Three QPRR models that meet the predictive capability criteria were developed. The relationships can be used to gain pharmacologically interesting information on the biologically active ampholytic substances.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is a typical practice nowadays to search for correlations between values of various measured physicochemical features of molecules or molecular systems and their predicted or measured structural or physicochemical characteristics [1,2,3,4,5,6,7]. Such correlations can be generally found using chemometric methods of analysis [8,9,10,11]. Following this trend, we applied in this work the Quantitative Property-Retention Relationship (QPRR) [12] approach for revealing possible relations between retention parameters of 40 substances exhibiting ampholytic properties owing to the presence of carboxylic, sulphonyl, sulphonamide, hydroxy, thio, amino, and imino groups or heterocyclic nitrogen atom (s) in their structures (Table 1S in the Supplementary materials)—obtained using HPLC with six polar and semipolar columns—and physicochemical (predicted) and spectral (experimental) characteristics of the compounds.
Since the analyzed compounds exhibit common property that is ampholyticity, but are significantly structurally diversified, we selected the abovementioned QPRR method, instead of more often applied QSRR (Quantitative Structure-Retention Relationship) approach [12,13,14,15,16]. We do so, having in mind that many drugs possessing similar structure exhibit different therapeutic features [17, 18]. On the other hand, similarities in the physicochemical properties of the compounds investigated should be reflected in their biological behavior.
The compounds investigated are natural and synthetic amino acids, pseudo-amino acids (containing an amino group in the chain or in a ring), sulphanilic acid, and its derivatives (sulphonamides) and derivatives of fluoroquinone and purine [17, 19, 20]. Among these compounds are precursors of biologically interesting peptides or proteins, others are important as vitamins or valuable pharmaceuticals. The choice of the compounds of a wide spectrum of biological activity and one common property (ampholyticity) for investigations gives a unique opportunity to reveal possible relations between retention parameters and physicochemical (predicted/experimental) descriptors, and afterwards to predict chromatographic parameters of untested substances, to gain valuable information on the biological activity/pharmacological relevance of ampholytic molecules and, to some extent, to model conditions of chromatographic separations.
Materials and methods
Chemicals
Data concerning the investigated compounds—their names, structure and properties—are compiled in the Supplementary materials (Table 1S). Detailed information on the sources of the target compounds is provided in our recent works [17, 19, 20].
Chromatographic investigations
Chromatographic analyses were carried out on a Waters SM 2690 Alliance HPLC system equipped with a PDA 996 spectroscopic diode detector and a Compaq Deckpro computer on which the Millennium 3.2 program for data collection and process control was installed. Six HPLC stainless steel columns were applied: Discovery HS PEG of 150 × 4 mm i.d. (Supelco) packed with silica gel chemically bounded with polyethylene glycol of particle size 5 μm (denoted further on as LKD); Hypersil HSA of 50 × 4.6 mm i.d. (Thermo-Hypersil-Keystone, Cheshire, UK) packed with silica gel bounded with human blood serum albumin of particle size 7 μm (denoted further on as LKH); IAM PC C10/C13 of 150 × 4.6 mm i.d. (Regis Chemical Company, Morton Grove, IL, USA) packed with silica gel chemically bounded through propylamine with phosphatidylcholine and subsequently endcapped through unreacted propylamine with methyl glycolate of particle size 12 μm (denoted further on as LKIAM); IC Pack Cation M/D of 150 × 3.9 mm i.d. (Waters) packed with silica gel chemically bounded with copolymer of butadiene and maleinic acid of particle size 5 μm (denoted further on as LKIC); Nucleosil 100-5 OH (Diol) of 250 × 4 mm i.d. (Macherey-Nagel, Düren, Germany) packed with silica gel chemically bounded with propylene glycol (propanediol) of particle size 5 μm (denoted further on as LKN); and Spheri-10 Anion AX-MP of 100 × 4.6. mm i.d. + 30 × 4.6. mm i.d. precolumn (Brownlee Laboratories, Santa Clara, CA, USA) packed with silica gel chemically bounded with polyethyleneimine of particle size 10 μm (denoted further on as LKS).
The compounds investigated were subjected to chromatographic analyses in isocratic conditions at ambient temperature (20 °C). The mobile phases were as follows: 0.025 M phosphate buffers of pH 2.5 and 7.0 (buffers of required pH were obtained by adding adequate portion of H3PO4 to 0.025 M aqueous solution of NaH2PO4) in the case of analyses carried out on the LKD column; acetonitrile 0.025 M phosphate buffers of pH 2.5 and 7.0 mixed in proportion 10:90 in the case of analyses carried out on the LKS, LKIC, and LKN columns or 20:80 in the case of analyses carried out on the LKIAM column; and 1-propanol:0.05 M phosphate buffer of pH 7.0 mixed in proportion 5:95 in the case of analyses carried out on the LKH column. All the mobile phases were filtered through a Whatman GF/F glass micro filter and degassed by ultrasonication immediately before use. The compounds investigated were dissolved in solvents present in mobile phases. The detection wavelength was 220 nm.
The HPLC retention factors (k) of the target compounds, equal to (tR − t0)/t0, were assessed on the basis of measured retention times (tR) and dead times (t0) by the Knox and Kaliszan method [21]. Since the amount of solvents in mobile phases never exceeded 20%, it was assumed that decadic logarithms of these factors, named subsequently retention parameters (logkw), correspond to pure buffers (subscript w indicates water). Thus, determined retention parameters, summarized in Table 2S (Supplementary materials), can be identified as logarithms of n-octanol/water partition coefficients [15]. The naming symbol of the retention parameter refers to the symbol of the column and round off value of buffer pH (e.g., logkwLKD2).
Physicochemical and spectral characteristics
Detailed information concerning the physicochemical and spectral characteristics of the investigated compounds used in chemometric analysis is given in Table 3S (Supplementary materials) [17, 19, 20]. The values of these characteristics are compiled in Table 4S (Supplementary materials).
Chemometric analysis
To find relations between determined retention parameters and physicochemical/spectral (predicted/measured) characteristics, the Quantitative Property-Retention Relationship (QPRR) approach was employed [17] for each of 11 experimental sets of data corresponding to six polar and semipolar columns with selected mobile phases and pH conditions. Following this approach, it was revealed that the GA-MLR method makes up a satisfactory tool for finding functional relations (models) between values of logkw and values of physicochemical/spectral characteristics (descriptors). In a result, three models relating the logkwLKD7, logkwLKIC2, and logkwLKN7 with the abovementioned descriptors were developed. Values of descriptors were, in each case, autoscaled. Genetic Algorithm (GA) was employed to select optimal set of descriptors [22]. The set of parameters applied to control GA was the size of population—100 and the mutation rate—45%. All chemometric calculations were done using QSARINS software [23]. The model fitting, robustness and predictive abilities were assessed by recommended procedures described elsewhere [24,25,26,27,28]. The applicability domain was evaluated by the Williams plot approach [29,30,31]. The Y-scrambling procedure was applied to confirm the model’s robustness.
Results and discussion
Resolving ability of HPLC columns
Unique collection of retention parameters (Table 2S in the Supplementary materials) provides an opportunity for evaluation of separating ability of columns used in HPLC analyses of ampholytic substances. A measure of the abovementioned feature can be the gap between logkw values for a given column and conditions. Values of this quantity compiled in Table 5S (Supplementary materials) reveal that the largest gap, and thus, the highest separating ability possess LKD, LKH, LKIC, and LKS columns, lower separating ability possesses LKIAM column and the lowest—LKN column. The above information forms a useful framework for planning chromatographic analyses and modeling conditions for separation of analyte components.
Relevance of retention parameters in the context of lipophilicity
Results of numerous investigations supported by OECD recommendations indicate that retention parameters (logkw) satisfactory approximate logarithms of n-octanol/water partition coefficients [32] which reflect lipophilicity of molecular systems, among other biologically active substances of pharmacological relevance. This important property determines features and behavior of biomolecules, e.g., ability to interact with proteins [32], and pharmacological and toxicological potency [33]. Retention parameters obtained in this work (Table 2S in the Supplementary materials) are a unique collection of data which carry information on lipophilicity of the ampholytic compounds investigated. This information refers to the certain pH values, 2.5 and 7.0, which correspond roughly to acidity of a medium in various parts of the alimentary canal, namely stomach (pH ~ 1–2) and intestine (pH ~ 6.8–7.4) [17]. The data collected can thus be helpful in explaining the pharmacological potency of the target compounds.
QPRR results
The GA-MLR methodology was employed to develop QPRR models allowing prediction of three chromatographic parameters: logkwLKD7, logkwLKIC2, and logkwLKN7 for ampholytic compounds based on their physicochemical characteristics.
Obtained models (equations) together with their statistical characteristics are presented in Table 1. Other details related to the models are summarized in Tables 6S–8S (Supplementary materials). The most optimal model developed for the LKD7 column is comprised of three descriptors: BE—binding energy (obtained with the AM1 method), TEAI—total energy (obtained at the ab initio level of theory), and Eabs (pH 7.0)—the energy of long wavelength absorption in buffer of pH 7.0. In the case of the LKIC2 column, the model is based on two descriptors: POL—polarizability (calculated with the AM1 method) and HE—hydration energy (obtained by QSAR approach). In the case of the third column, the lipophilicity (logkwLKN7) is defined as a function of three descriptors: OSMAX—the oscillator strength corresponding to the first long wavelength electronic transition (calculated by AM1/CI), POLAI—polarizability (calculated with HF method), and Eabs (pH 7.0)—the energy of long wavelength absorption in buffer of pH 7.0.
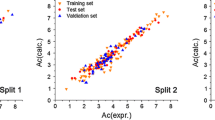
High values of R2, Q2CV, and Q2Ext and relatively low values of errors represented by RMSEC, RMSECV, RMSEP confirm satisfactory the goodness-of-fit, robustness, and predictive ability of the developed QPRR models. The quality of the models is additionally proved by the visual correlation between the observed and predicted values of retention parameters for the training (T) and validation (V) sets (Fig. 1). Scrambling procedure confirms that models are not correlated by chance (Fig. 1S in the Supplementary materials).
Calculated versus observed values for models summarized in Table 1
Williams plots were generated to verify the applicability domain of the developed models (Fig. 2). According to this approach, if residuals do not differ by ± 3 standard deviations from the mean value and are similar for the sets of training and validation compounds (this similarity is expressed by so-called leverages), then the models (equations) can be used to predict logkw of untested compounds. Since no outlying predictions are observed in Fig. 2 one can conclude that the developed models present satisfactory predictive capability.
Williams plot: standardized residuals versus leverages. Solid lines indicate ± 3 standard deviation units, dashed line indicates the threshold value for models summarized in Table 1
Descriptors relevance
All presented models reflect the influence of physicochemical/spectral characteristics on the lipophilicity defined as logkw. The models for LKD7 and LKN7 columns relate negatively respective logkw values with spectral characteristics: Eabs (pH 7.0) depending on the pH (in both cases) and OSMAX (in the latter case), namely values of the logkw increase with the decrease of values of these latter descriptors. Physicochemical descriptors POL and POLAI representing polarizability of molecules, which influence positively logkw values for LKIC and LKN columns, respectively, are known to have an impact on the overall hydrophobic properties of molecules [34]. According to the developed models, an increase of polarizability of molecules causes an increase of their lipophilicity, which is well-reflected in the data, e.g., flamequine, theobromine, and L-levodopa. The model for LKIC2 relates positively relevant logkw values with hydration energy (HE), reflecting the energy released in a solvation process [35]. Thus, compounds exhibiting a higher tendency to hydration, such as L-methionine, piroxicam or p-aminophenol, should be more lipophilic. Total energy (TEAI) and binding energy (BE), which influence logkwLKD7 values, are a measure of stability of molecules and to some extent their susceptibility to interact with molecules from surroundings, e.g., water molecules. According to the developed equation, more stable molecules are more lipophilic (and more hydrophobic), e.g., hydrochlorothiazole, sulfathiazole, and sulfanilamide. The above discussion demonstrates that found dependencies of logkw on physicochemical/spectral descriptors can be a useful tool for predicting of features and behavior of unknown ampholytic compounds.
Concluding remarks
On the basis of the QPRR approach, three models relating the logkw values with physicochemical (predicted/spectral) parameters of ampholytic biologically active substances of pharmacological relevance were developed using GA-MLR method. These models concern three chromatographic columns, among six selected for investigations, and three variants of experimental conditions, among 11 considered. From this information emerges that the occurrence of the correlation between the abovementioned quantities is not a common case.
The QPRR models form a useful platform for predicting retention parameters of untested compounds having common features (e.g., ampholyticity) with the tested compounds. This approach, utilizing physicochemical characteristics of the compounds as chemometric descriptors, instead of structural ones used in QSRR models, allows extending the applicability of the developed models for predicting retention parameters of structurally diversified substances.
The relationships found can be used to gain pharmacologically interesting information on the biologically active ampholytic substances. The log kw values determined in this work are a unique collection of chromatographic parameters which when used as a measure of lipophilicity can be very helpful in assessing pharmacological potency of the compounds investigated.
References
C. Hansch, A. Leo, Exploring QSAR. Fundamentals and applications in chemistry and biology, American Chemical Society, Washington DC, 1995
Markuszewski M, Kaliszan R (2002) Quantitative structure-retention relationships in affinity high-performance liquid chromatography. J Chromatogr B 768:55–66
Baczek T, Kaliszan R (2001) Quantitative structure/retention relationships in affinity chromatography. J Biochem Bioph Meth 49:83–98
Kaliszan R (1992) Quantitative structure-retention relationships. Anal Chem 64:A619–A631
Kaliszan R (2007) QSRR: quantitative structure-(chromatographic) retention relationships. Chem Rev 107:3212–3246
Kaliszan R (2013) Quantitative structure property (retention) relationships in liquid chromatography. Liquid Chromatography: Fundamentals Instrumentation:385–405
Gorynski K, Bojko B, Nowaczyk A, Bucinski A, Pawliszyn J, Kaliszan R (2013) Quantitative structure-retention relationships models for prediction of high performance liquid chromatography retention time of small molecules: endogenous metabolites and banned compounds. Anal Chim Acta 797:13–19
Roy K, Kar S, Das RN (2015) A primer on QSAR/QSPR modeling: fundamental concepts. Springer, London
Bielinska-Waz D, Waz P, Jagiello K, Puzyn T (2014) Spectral density distribution moments as novel descriptors for QSAR/QSPR. Struct Chem 25:29–35
Jagiello K, Sosnowska A, Walker S, Haranczyk M, Gajewicz A, Kawai T, Suzuki N, Leszczynski J, Puzyn T (2014) Direct QSPR: the most efficient way of predicting organic carbon/water partition coefficient (log K (OC)) for polyhalogenated POPs. Struct Chem 25:997–1004
Sosnowska A, Barycki M, Jagiello K, Haranczyk M, Gajewicz A, Kawai T, Suzuki N, Puzyn T (2014) Predicting enthalpy of vaporization for persistent organic pollutants with quantitative structure-property relationship (QSPR) incorporating the influence of temperature on volatility. Atmos Environ 87:10–18
Heberger K (2007) Quantitative structure-(chromatographic) retention relationships. J Chromatogr A 1158:273–305
Kaliszan R, van Straten MA, Markuszewski M, Cramers CA, Claessens HA (1999) Molecular mechanism of retention in reversed-phase high-performance liquid chromatography and classification of modern stationary phases by using quantitative structure-retention relationships. J Chromatogr A 855:455–486
Al-Haj MA, Kaliszan R, Nasal A (1999) Test analytes for studies of the molecular mechanism of chromatographic separations by quantitative structure-retention relationships. Anal Chem 71:2976–2985
Braumann T, Weber G, Grimme LH (1983) Quantitative structure-activity-relationships for herbicides - reversed-phase liquid-chromatographic retention parameter, log kw, versus liquid-liquid partition-coefficient as a model of the hydrophobicity of phenylureas, s-triazines and phenoxycarbonic acid-derivatives. J Chromatogr 261:329–343
Flieger J, Swieboda R, Tatarczak M (2007) Chemometric analysis of retention data from salting-out thin-layer chromatography in relation to structural parameters and biological activity of chosen sulphonamides. J Chromatogr B 846:334–340
U. Judycka, K. Jagiello, M. Gromelski, L. Bober, J. Blazejowski, T. Puzyn, Chemometric approach to correlations between retention parameters of non-polar HPLC columns and physicochemical characteristics for ampholytic subtances of biological and pharmaceutical relevance, J Chromatogr B, after review (minor revision in progress)
Jagiello K, Mostrag-Szlichtyng A, Gajewicz A, Kawai T, Imaizumi Y, Sakurai T, Yamamoto H, Tatarazako N, Mizukawa K, Aoki Y, Suzuki N, Watanabe H, Puzyn T (2015) Towards modelling of the environmental fate of pharmaceuticals using the QSPR-MM scheme. Environ Model Softw 72:147–154
Judycka U, Jagiello K, Bober L, Blazejowski J, Puzyn T (2018) Assessing therepeutic relevance of biologically interesting, ampholytic subtances based on their physicochemical and spectral characteristics with chemometric tools. Chem Phys Lett 701:58–64
Judycka-Proma U, Bober L, Gajewicz A, Puzyn T, Blazejowski J (2015) Chemometric analysis of correlations between electronic absorption characteristics and structural and/or physicochemical parameters for ampholytic substances of biological and pharmaceutical relevance. Spectrochim Acta A Mol Biomol Spectrosc 138:700–710
Knox JH, Kaliszan R (1985) Theory of solvent disturbance peaks and experimental-determination of thermodynamic dead-volume in column liquid-chromatography. J Chromatogr 349:211–234
Cho SJ, Hermsmeier MA (2002) Genetic algorithm guided selection: variable selection and subset selection. J Chem Inf Comp Sci 42:927–936
Gramatica P, Chirico N, Papa E, Cassani S, Kovarich S (2013) QSARINS: a new software for the development, analysis, and validation of QSAR MLR models. J Comput Chem 34:2121–2132
Gramatica P (2013) On the development and validation of QSAR models. Methods Mol Biol 930:499–526
Chirico N, Gramatica P (2011) Real external Predictivity of QSAR models: how to evaluate it? Comparison of different validation criteria and proposal of using the concordance correlation coefficient. J Chem Inf Model 51:2320–2335
Chirico N, Gramatica P (2012) Real external predictivity of QSAR models. Part 2. New intercomparable thresholds for different validation criteria and the need for scatter plot inspection. J Chem Inf Model 52:2044–2058
Roy K, Mitra I (2011) On various metrics used for validation of predictive QSAR models with applications in virtual screening and focused library design. Comb Chem High T Scr 14:450–474
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring r (m)(2) metrics for validation of QSPR models. Chemometr Intell Lab 107:194–205
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) QSAR applicability domain estimation by projection of the training set in descriptor space: a review. Atla-Altern Lab Anim 33:445–459
Nikolova-Jeliazkova N, Jaworska J (2005) An approach to determining applicability domains for QSAR group contribution models: an analysis of SRC KOWWIN. Atla-Altern Lab Anim 33:461–470
Netzeva TI, Worth AP, Aldenberg T, Benigni R, Cronin MTD, Gramatica P, Jaworska JS, Kahn S, Klopman G, Marchant CA, Myatt G, Nikolova-Jeliazkova N, Patlewicz GY, Perkins R, Roberts DW, Schultz TW, Stanton DT, van de Sandt JJM, Tong WD, Veith G, Yang CH (2005) Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships - the report and recommendations of ECVAM Workshop 52. Atla-Altern Lab Anim 33:155–173
Nasal A, Siluk D, Kaliszan R (2003) Chromatographic retention parameters in medicinal chemistry and molecular pharmacology. Curr Med Chem 10:381–426
Koba M, Baczek T, Marszall MP (2012) Importance of retention data from affinity and reverse-phase high-performance liquid chromatography on antitumor activity prediction of imidazoacridinones using QSAR strategy. J Pharm Biomed Anal 64-65:87–93
Lewis DFV (1989) The calculation of molar polarizabilities by the CNDO/2 method - correlation with the hydrophobic parameter. Log-P, J Comput Chem 10:145–151
Paterson Y, Nemethy G, Scheraga HA (1982) An empirical potential function for the interaction between univalent ions in water. J Solut Chem 11:831–856
Funding
The authors thank the Polish Ministry of Science and Higher Education (Grant No. DS 530-8637-D510-17) and Polpharma S.A. for financial support of investigations.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOC 1187 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Judycka, U., Jagiello, K., Gromelski, M. et al. Chemometric outlook on correlations between retention parameters of polar and semipolar HPLC columns and physicochemical characteristics of ampholytic substances of biological and pharmaceutical relevance. Struct Chem 29, 1839–1844 (2018). https://doi.org/10.1007/s11224-018-1174-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-018-1174-5