Abstract
Diclofenac is the world known nonsteroidal anti-inflammatory drug (NSAID) predicted before its syntesis on the basis of the model COX enzyme. Due to its specific structural properties the drug possesses high reactivity and outstanding tolerability. Among the key features defining the diclofenac structure is intramolecular N-H ⋯O hydrogen bond confirmed during the X-ray analysis. In the present research we use static DFT calculations, the Quantum Theory of Atoms in Molecules and non-covalent interactions (NCI) index to confirm the additional intramolecular interactions, which influence the drug molecular structure. We focus on the structural stability of diclofenac as the result of the hydrogen bonds breaking/formation at finite temperature utilizing ab initio molecular dynamics simulations. The lifetime of different intramolecular hydrogen bonds is estimated. We perform also the comparative analysis of the structural stability of ibuprofen and ketoprofen molecules in the gas phase at 300 K with respect to diclofenac in terms of the NSAID inhibition activity. Due to the detailed description of diclofenac intramolecular interactions, possible drug modifications for its enhanced water solubility can be suggested.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the most widely prescribed nonsteroidal anti-inflammatory drug (NSAID) products, diclofenac (DCL) ranks at the first place [1]. It is known as an effective cyclooxygenase (COX) inhibitor with anti-inflammatory, analgetic and antipyretic properties [2] with the best inhibition activity of the COX-2 enzyme [3], comparing with other NSAIDs, non-selective to this enzyme. Recently, DCL treatment was found to have even an anti-cancer effects [4].
The history of diclofenac (DCL) started at early 70s when, on the basis of both experimental and clinical findings, the features of an ideal nonsteroidal anti-inflammatory drug were postulated [5–8]. They said that an effective antirheumatic agent should have an acidity constant between 4 and 5, a partition coefficient (octanol/water at pH 7.4) of c.a. 10, and two aromatic rings maximally twisted in relation to each other [8]. Several factors were taken into account during the mentioned analysis: (i) drug transport through biologic membranes, (ii) the atomic and spatial structure of the molecule to fit the COX receptor, and (iii) the electronic structure, which controls the specific interaction between the drug and the receptor. Among more than 200 NSAID agents tested [8], including 36 congeners of diclofenac [9], DCL (with an acidity constant of 4.0 and a partition coefficient of 13.4) was found to possess the most interesting pharmacologic properties caused as the result of the specific structure.
DCL molecule consists of two aromatic rings: the phenylacetic and chlorine functionalized, and the secondary amino group, which is their linkage (see Fig. 1). As the result of the repulsion interations between the acetate group and the Cl atoms, the aromatic rings are twisted with respect to each other (the angle of twist is about 69 ° [8]), what enables the DCL specific interactions with the COX enzyme [7]. Moreover, such arrangement of the DCL phenyl rings provide a good fit in the substrate-binding pocket of the COX increasing its inhibition activity [5, 10]. Apart from the mentioned DCL structural feature, its secondary amino group participates in a bifurcated intramolecular hydrogen bond, interacting with the carbonyl oxygen and the chlorine atoms [9, 11]. The existence of N-H ⋯Cl H-bond was confirmed only in the X-ray analysis [11] and this weak type of interactions is rarely taken into consideration during the experimental investigation of DCL. In contrast, the N-H ⋯O H-bond is mentioned as characteristic intramolecular DCL hydrogen bond [8, 9, 12]. N-H ⋯O attractive interaction is known as strong (resonance-assisted) hydrogen bond [13]. The structural parameters of intramolecular N-H ⋯O H-bond for β-ketoarylhydrazones are in the range of 1.76–2.32 Å and 99–140 ° for the H ⋯O distance and NHO angle, respectively [14]. Simultaneously, the energy of this H-bond can equal even 60 kJ/mol.
Since, similarly to other NSAID, diclofenac therapy causes gastrointestinal, cardiovascular and renal adverse effects [15, 16], a lot of different DCL-containing drug products are developed [17] to improve the drug efficacy, tolerability and patient convenience. The existence of the mentioned intramolecular N-H ⋯O H-bond in DCL is utilized to modify the drug with, e.g., transition metals [18–20], cyclodextrin [21] or PEG [22], to increase its water solubility and oral bioavailability. Up to now, no research on the N-H ⋯O dynamics at finite temperature was made. Since this H-bond is crucial either during DCL modification or in the COX inhibion, we are focused on its detailed analysis utilizing both static DFT calculations and Car–Parrinello molecular dynamics simulations.
The DCL specific structure is caused not only by N-H ⋯O H-bond, but also by other intramolecular interactions, e.g. N-H ⋯Cl, O-H ⋯N H-bonds, vdW interactions and steric clashes. Their mutual interplay defines the drug structural stability and inhibition activity. As it was not studied before, in the present work the systematic study on the DCL structural rearrangements and intramolecular interactions at 300 K are investigated with the direct comparison with other known NSAIDs: ketoprofen and ibuprofen (see Fig. 1). The results obtained might be successfully used during the detailed analysis of the possible DCL modifications with saving its specific bioactivity. Taking into account high computational cost of an aqueous environment in the present simulations, we use only gas-phase calculations to establish the role of intramolecular interactions on the DCL structural stability.
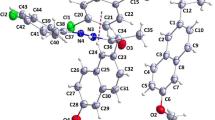
Optimized global minimum A (a) and selected local minima B (b) and C (c) of the diclofenac molecule obtained using DFT static calculations. The NCI surfaces correspond to s = 0.4 a.u. and a color scale of -0.05 <ρ< 0.05 a.u. The H-bonds (confirmed by QTAIM) and the C-C-N-C diheral angle are marked in black dash lines and red circles, respectively
Computational methods
The main part of the studies performed focuses on the analysis of the structural stability of DCL molecule using Car–Parrinello molecular dynamics simulations [23]. Before it, the geometry optimization of the diclofenac molecule was performed using density functional theory (DFT) approach with B3LYP functional and localized basis set 6-31G(d,p) using Gaussian09 program package [24]. Among 21 calculated local minima, three DCL structures (see Fig. 2) were chosen as the initial atomic configurations for the molecular dynamics (MD) simulations of isolated DCL molecules in the gas phase. The first structure (A) is the most stable DCL structure in the gas phase, the second one (B) is similar to the DCL crystal structure [11], while the third one (C) is the most unstable structure of diclofenac. MD simulations were performed according to the Car–Parrinello scheme [23] using the CPMD program package (version 4.1.0) [25] with the general empirical dispersion correction (D2) of Stefan Grimme [26]. All calculations were made utilizing Perdew-Burke-Ernzerhof (PBE) exchange-correlation functional and the plane-wave (PW) basis set with the kinetic energy cutoff of 30 Ry. The PBE functional was previously shown to give the satisfactory precision in the description of systems with hydrogen bonding [27]. The effective potential of ions was described by Vanderbilt ultrasoft pseudopotentials [28]. We used the unit cell of 22.0Å ⋅22.0Å ⋅22.0Å for periodic boundary conditions to prevent the interactions between neighbor DCL molecules. The Poisson equation for isolated system was solved using Martyna–Tuckerman method [29]. All wavefunctions were fully converged (1 ⋅10−8 a.u.) and the electronic degrees of freedom were quenched to the Born–Oppenheimer surface before starting the simulations. We used Nosé–Hoover [30, 31] chain thermostats on ions and electrons in the canonical ensemble to keep the adiabatic isolation between the fictitious electron and nuclear dynamics. The frequency for the ionic thermostat was set to 1500 cm −1, while for the electron thermostat on 10000 cm −1. All simulations were 30 ps long with the preceded equilibration procedure of 10 ps. The time step and the fictitious electronic mass of MD simulations were set to 4 a.u. and 400 a.u., respectively. The simulations were performed at finite temperature of 300 K.
The systematic description of intramolecular hydrogen bonds and other noncovalent interactions in diclofenac was performed using the Quantum Theory of Atoms in Molecules (QTAIM) [32] and the non-covalent interactions (NCI) index [33, 34]. For this purpose AIMALL program (version 13.05.06) [35] and NCIPLOT program (version 3.0) [36] were utilized. The fully converged wavefunctions of the optimized DCL molecules were generated utilizing the hybrid PBE functional with empirical dispersion correction D2 and the localized basis set (aug-cc-pVDZ) using Gaussian09 package [24]. The QTAIM was used to confirm quantitatively the existence of hydrogen bonds in DCL by the analysis of the electron density and its Laplacian at the bond critical points (BCP). The aim of the NCI index was to detect qualitively and vizualize hydrogen bonds, vdW interactions and steric clashes in the molecule on the basis of the electron densities and their reduced gradients. The vizualization of the results obtained were made using VMD program package [37].
The same computational setup was used to analyze ketoprofen and ibuprofen molecules. Only the frequencies of the thermostats were set to the other values: 1260 cm −1 and 10000 cm −1 for ketoprofen; 3000 cm −1 and 15000 cm −1 for ibuprofen, on the ionic and electron thermostats, respectively.
Results and discussion
Static calculations
In the literature, a lot of investigations concerning the diclofenac interactions with biomolecules, drug modifiers etc. are present, but only the crystal structure of the DCL molecule is known [11]. As the result, before the analysis of the DCL structural stability we have predicted the most probable drug structure in the gas phase. Figure 2 shows three of 21 calculated DCL conformers with the corresponding value of their relative energy. We have chosen only three DCL structures to show the significant structural differences of different DCL conformers at 300 K using molecular dynamics simulations (see Section “Molecular dynamics simulations”). It should be noted that the examined structure B represents the DCL crystal structure, reported in Ref. [11]. The only difference between these DCL structures is the C-C-N-C dihedral angle, which equals to 60.47 ° for the calculated structure and 62.60 °/62.19 ° for HD1/HD2 polymorphic DCL forms.
For the best understanding of the influence of the noncovalent interactions (NCI) on the defining the drug structure, the NCI surfaces are shown simultaneously in Fig. 2. To analyze intramolecular hydrogen bonds in the molecule we have used two the most popular theoretical techniques: the Bader analysis [32] and the noncovalent (NCI) index [33]. The first one uses the value of the electron density and its Laplacian at the bond critical points (BCP) to estimate the hydrogen bond existence and strength – according to U. Koch and P. Popelier [38], the hydrogen bond exists if the electron density is in the range of 0.002–0.035 a.u. and its Laplacian – 0.024–0.139 a.u. The second one provides three-dimensional regions around BCPs on the basis of the sign of second eigenvalue (λ 2) of the electron-density Hessian matrix [34] to vizualize both attractive and repulsive interactions in the real-space. In general, NCI surfaces with density values in the range of 0.005 < ρ < 0.05 a.u. characterize strong noncovalent interactions, including H-bonds (negative λ 2) and steric clashes (positive λ 2). Characteristic densities of vdW interactions are smaller than H-bonds. Surfaces with very low density values (ρ < 0.005 a.u.) shows the existence of weak dispersion interactions.
From Fig. 2 one can see that the noncovalent interactions pattern for the DCL conformers is similar. All structures are characterized by three types of the NCI interactions: hydrogen bonds, vdW interactions and steric clashes, which are mapped in green. For the best analysis of the NCI index, the relationship between reduced density gradient and the sign of λ 2 is shown in Fig. 3. The DCL global minimum (structure A) possesses two intramolecular hydrogen bonds: N-H ⋯O and N-H ⋯Cl, with the H ⋯B distance of 2.02 Å and 2.56 Å, respectively. Because of the low linearity of the N-H ⋯Cl hydrogen bond (104.9 °), only N-H ⋯O H-bond, with the electron density value of 0.0231 a.u. and its Laplacian of 0.0673 a.u., was identified using the QTAIM analysis (see Table 1). As it was suggested in Ref. [8], the DCL structure is influenced by the repulsion between the aromatic rings. This fact is also confirmed here due to the NCI analysis; the repulsion interaction is shown as large steric clash (in green) in Fig. 2. The strength of these interactions is comparable to the attractive ones, caused by the hydrogen bonds formation (see the values of the reduced electron density gradients (s) in Fig. 3). As the result, the angle of the twist between the DCL phenyl rings, measured by the value of the C-C-N-C dihedral angle (marked in red circles), is equal to 58.55 °. The structure A is the most stable one, as the result its relative energy value equals to 0 kcal/mol. The relative energy values for other DCL local minima were calculated with respect to this structure.
NCI analysis of diclofenac local minima (A, B, C): the relationship between reduced density gradient (s) and the sign of the second eigenvalue (λ 2) of the electron-density Hessian. Peaks appear at ρ≃ –0.05 a.u., ρ≃±0.01 a.u. and ρ≃ 0.05 a.u. characterize H-bonds, vdW interactions, and steric clashes, respectively
The structure B of the diclofenac molecule is very similar to the global minimum structure. The pattern of the NCI interactions is almost the same, e.g., the structural and topological parameters of the hydrogen bonds (Table 1) or NCI parameters (Fig. 3). The only difference between these two structures is the geometry of the COOH group with respect to the CH 2 fragment and the mutual directionality of the DCL phenyl rings. It can be easily seen in Fig. 4, where the superposition of the structures A and B is shown. Such small structural differences cause the change of the repulsive wall between the phenyl rings, making the DCL molecule less stable (it was confirmed during ab initio molecular dynamics simulations, see Section “Molecular dynamics simulations”), therefore, the absolute energies of these structures differ by 6.05 kcal/mol. Additionally, these differences cause that the DCL structure B is able to inhibit COX-2 receptor, because such mutual phenyl ring position of DCL is similar to the COX-2 substate [39]. Therefore, diclofenac structure bounded to the active site of the enzyme is similar to the mentioned structure B.
Completely different conformation of the diclofenac molecule represents the least stable structure (marked as C in Fig. 2), with the relative energy value of 13.19 kcal/mol. The superposition between the structures A and C is shown in Fig. 4. In comparison to the global minimum, in the structure C there is no evidence for the formation of the intramolecular N-H ⋯O hydrogen bond, which is the most characteristic and energetically stable in the case of DCL molecule. Instead of the strong N-H ⋯O H-bond, the weak C-H ⋯O hydrogen bond with the H ⋯O distance and CHO angle of 2.45 Å and 106.2 °, respectively (see Table 1), is formed. The strength of the repulsion wall between the DCL phenyl rings is slightly weaker than in the case of the structures A and B (see the values of the electron density with positive sign of λ 2 in Fig. 3). As the result, the angle of the twist between the phenyl rings is smaller and equals 30.87 °. There is an extra repulsion between the phenylacetic ring and the secondary amino group, based on the hydrogen-hydrogen repulsion interactions (see Fig. 2). The structure C, as A and B, possesses N-H ⋯Cl H-bond, but due to its slightly higher linearity (111.1 °), its existence was confirmed by the QTAIM analysis (see Table 1).
Molecular dynamics simulations
The best way to estimate the structural stability of diclofenac molecule is to analyze its dynamics using ab initio molecular dynamics (MD) simulations at finite temperature. In the present work, MD simulations were performed for all previously analyzed DCL conformers (see Section “Static calculations”) to provide a detailed description of possible DCL structural rearrangements in the gas phase at 300 K. The selected snapshots illustrating the DCL conformation at finite time of the simulation are shown in Fig. 5. As it was mentioned before, the molecular structure of diclofenac depends on the repulsive character of the phenyl rings, which cause the specific twist between them and enable the formation of additional intramolecular hydrogen bonds stabilizing the particular DCL conformation. Consequently, we are analizing DCL hydrogen bonds breaking and formation to estimate their impact on the DCL structural rearrangements, which can be described through the changes of the C-C-N-C dihedral angle (see Section “Static calculations”).
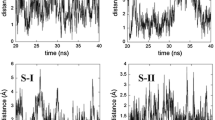
The DCL global minimum structure (A) is characterized with the highest structural stability at 300 K, which is depicted in the corresponding snapshots in Fig. 5. It can be confirmed by the analysis of the time evolution of its C-C-N-C dihedral angle (black curve in Fig. 6), which is relatively stable during the simulation time. The values of the C-C-N-C dihedral angle oscillate around 60 °, what is consistent with its value (69 ° [8] and 62 ° [11]) obtained in the X-ray analysis. Such DCL conformation is similar to the one obtained using static calculations (structure A), where two hydrogen intramolecular bonds are formed: N-H ⋯O and N-H ⋯Cl. The existence of both H-bonds can be confirmed on the basis of H ⋯B distance and AHB angle, where A-H and B are H-bond donor and acceptor, respectively. According to the latest IUPAC definition [40]: the closer the H-bond angle is to 180 °, the stronger is the hydrogen bond and the shorter is the H ⋯B distance. Moreover, the preferable value for the AHB angle is more than 110 °. Fig. 7 represents changes of the H ⋯B distance and the AHB angle of N-H ⋯O and N-H ⋯Cl hydrogen bonds during the MD simulation. The analysis of the radial distribution function of H ⋯B distance indicates the average H ⋯O distance of 2.08 Å and H ⋯Cl – 2.56 Å. Both hydrogen bonds are stable during the simulation time. N-H ⋯O hydrogen bond is stronger than N-H ⋯Cl one.
Even if the first DCL local minimum (structure B) slightly differs from the structure A in static calculations, its structural stability at 300 K is definitely different (see Fig. 5). Apart from the characteristic for diclofenac N-H ⋯O and N-H ⋯Cl hydrogen bonds, there are structural rearrangements caused by the O-H ⋯N H-bond formation (snapshot “1” in Fig. 5) with the maximum lifetime of 1.5 ps (Fig. 8). This hydrogen bond defines the DCL conformation during the first 6 ps (see Fig. 6), while no N-H ⋯O and N-H ⋯Cl hydrogen bonds are formed. Such formed structure is not stable, therefore, it changes to the one depicted in the snapshot “2” in Fig. 5, where the C-C-N-C dihedral angle equals to c.a. 180 °. The lifetime of such DCL conformation, without hydrogen bonds formed, is only 1 ps (Fig. 6). Since c.a. 8 ps DCL rearranges to the structure shown in the snapshot “3”, which reminds the structure B in the static calculations. The time evolution of the C-C-N-C dihedral angle of such DCL conformation (Fig. 6) shows its relatively high structural stability till the end of the MD simulation. This structure is defined by intramolecular N-H ⋯O and N-H ⋯Cl hydrogen bonds (see black and red curves in Fig. 8) with the average H ⋯B distance of 2.06 Å and 2.48 Å, respectively. Taking into account the COX-2 structure and DCL binding to its active site [39], the structure B is similar to the DCL structure, which participates in the inhibition process of the COX-2. Due to higher flexibility of the N-H ⋯O H-bond in the structure B, comparing to the structure A (see Fig. 10), such DCL conformation can interact with the COX-2 active site via hydrogen bonds between DCL carboxyl group and Ser-530/Tyr-385.
The least stable structure in the static calculations (structure C) is very stable during the first 16 ps of MD simulation (see Fig. 6). The C-C-N-C dihedral angle, indicating the twist between the phenyl rings, is close to the one calculated in Section “Static calculations”. Such structure formed possesses stable intramolecular N-H ⋯Cl hydrogen bond with the average H ⋯Cl distance of 2.43 Å. Since c.a. 12 ps of the simulation, temporary O-H ⋯N H-bonds with maximum lifetime of 1.5 ps are formed (see Fig. 9 and the snapshot “2” in Fig. 5). Despite the relatively high structural stability of the analyzed DCL conformation, it changes the character of its rearrangements, which starts to remind dynamics of the structure B during it first 6 ps of the simulation (see Fig. 6). Such newly formed conformation is stable for c.a. 4 ps. Later, the structure with N-H ⋯O and N-H ⋯Cl hydrogen bonds is formed (see the snapshot “3” in Fig. 5), which after 2 ps rearranges again to unstable DCL conformation. Taking into account similar character of the rearrangements of the structure C (after 16 ps) and the structure B (until 6 ps), one can predict that in the finite time of the simulation the structure C transforms to B, which differs with the global minimum structure mainly by the direction of the twist between the DCL phenyl rings.
The plot of the time evolution of the H ⋯B distance in N-H ⋯O H-bond of all examined DCL conformations is shown in Fig. 10. This intramolecular hydrogen bond is considered, in the literature, as the most stable and characteristic DCL hydrogen bond [8]. Figure 10 indicates that N-H ⋯O H-bond is not stable in all DCL conformations, thus, not only this H-bond should be taken into consideration during diclofenac modifications. The highest stability of this H-bond is seen only in the case of the global minimum structure of DCL.
Comparison of NSAID inhibition activity
Most of chemical and biochemical reactions between a protein and a drug are controlled by noncovalent interactions, which are dependent on a drug molecular structure. According to results reported in Ref. [39, 41], depending on the NSAID structure, different junctions to the COX-2 active site for the inhibitor binding are possible. Most of the NSAID molecules inhibit COX-2 through the carboxyl group coordination with Arg-120 and Tyr-355 near the membrane interface, while the selective COX-2 inhibitors bind to the enzyme side pocket [39]. Diclofenac shows distinct pharmacophore behavior [42]. Due to its specific structure, DCL binds to COX-2 similarly to the non-productive arachidonic acid (the target substate of COX-2) through the hydrogen bond with Tyr-385 and Ser-530 [39]. Therefore, its COX-2 inhibition activity was reported to be the best [3]. The detailed structure of the COX-2 enzyme with DCL bound to the active site is a supplementary material to Ref. [39] and can be found in PDB database under the name of 1PXX. The structure of DCL bounded to COX-2 is similar to the crystal DCL structure, reported in the present work as the structure B.
Ibuprofen and ketoprofen are also widely known NSAID with similar to diclofenac acidity constants: 4.5 and 4.0, respectively. Moreover, ibuprofen has even similar partition coefficient of 12.7. But, among these three NSAID representatives, only DCL contains phenyl rings twisted with respect to each other, which enables DCL to possess similar geometry to the COX-2 substrate. As a result, ketoprofen and ibuprofen take the eighth and the fourteenth place on the COX-2 inhibition activity list, mentioned in Ref. [3]. The same trend is present in the case of piroxicam and phenylbutazone [8].
To compare the structural differences between diclofenac, ketoprofen and ibuprofen in detail, their global minimum structures with the NCI surfaces are shown in Fig. 11. It can be seen that ketoprofen has similar to DCL structural features, but instead of the chlorinated phenyl ring, the phenyl ring without substituents is present, and secondary amino group is changed to carbonyl one. Due to such changes, there is weaker repulsion wall and lower angle of a twist between the phenyl rings. Only weak intramolecular C-H ⋯O hydrogen bonds can be formed. The existence of the C-H ⋯O H-bond can be confirmed due to the NCI index (see lower chart in Fig. 11). The QTAIM and NCI analysis of the ibuprofen molecule (structure (c) in Fig. 11) indicate that there is no specific repulsion in the molecule and no evidence for intramolecular hydrogen bonds formation. At the same time, the value of the C-C-C-C dihedral angle is close to the C-C-N-C observed in DCL. The structural stability of all these NSAID molecules was analyzed using ab initio MD at 300 K. The results obtained showed no significant structural changes in ibuprofen molecule and higher mobility of the phenyl ring s in ketoprofen, comparing with DCL. The higher rotational flexibility of ketoprofen might unable its bonding with Ser-530, which interacts efficiently with DCL. The similar dependence was reported by Llorens et al. [42] for fenoprofen. Therefore, we suggest that the higher inhibition activity of diclofenac can be explained on the basis of the existence of intramolecular hydrogen bonds, which stabilize significantly the drug structure in its preferable, for COX-2 enzyme, configuration with maximum twist between the phenyl rings.
Optimized global minima of diclofenac (a), ketoprofen (b) and ibuprofen (c) molecules obtained using DFT static calculations and the corresponding s(ρ) plots for their SCF density. The NCI surfaces correspond to s = 0.4 a.u. and a color scale of -0.04 <ρ< 0.04 a.u. The H-bonds (confirmed by QTAIM) and C-C-N-C diheral angle are marked in black and red dash lines, respectively
Conclusions
We used static DFT calculations and ab initio molecular dynamics simulations at 300 K to analyze the structural stability of the diclofenac molecule and estimate its influence on the drug inhibition activity. Noncovalent interactions, which define the molecular structure of diclofenac, were identified using the Quantum Theory of Atoms in Molecules and the NCI index. The existence of intramolecular hydrogen bonds in diclofenac: N-H ⋯O, N-H ⋯Cl and O-H ⋯N, was also confirmed. The time-resolved properties of the most important structural descriptors were presented and discussed in detail.
The most probable molecular conformation of diclofenac in the gas phase is the global minimum structure, which is characterized with high structural stability at 300 K and is stabilized by intramolecular N-H ⋯O and N-H ⋯Cl hydrogen bonds. However, the DCL global minimum has not been found during the inhibition process of COX-2 as the result of the changed mutual orientation of the phenyl rings, which unable the hydrogen bond formation with Tyr-385 and Ser-530. The structure B, which has been found to bind to the enzyme active site, is less stable than global minimum A and represents the crystal DCL structure. It possesses higher flexibility of the DCL characteristic N-H ⋯O hydrogen bond, enabling easier coordination of the COX-2 active site. The lifetime of temporary O-H ⋯N hydrogen bond, which determines some less stable drug rearrangements, is maximum 1.5 ps.
The direct comparison of the structural stability of diclofenac and other NSAID molecules (ketoprofen and ibuprofen) indicate that diclofenac possesses higher COX-2 inhibition activity as the result of effective structure stabilization with intramolecular hydrogen bonds formation and characteristic repulsion wall enabling the maximum twist between the drug phenyl rings. These noncovalent interactions ensure the DCL molecule to have geometry similar to the non-productive arachidonic acid, which is the substrate of COX-2. Thus, diclofenac inhibits COX-2 in the orientation that prevent interactions with Arg-120, which are present during the COX-2 inhibition by other NSAID.
References
McGettigan P, Henry D (2013) Use of non-steroidal anti-inflammatory drugs that elevate cardiovascular risk: an examination of sales and essential medicines lists in low-, middle-, and high-income countries. PLoS Med 10 (2):e1001388
Schwartz JI, Dallob AL, Larson PJ, Laterza OF, Miller J, Royalty J, Snyder KM, Chappell DL, Hilliard DA, Flynn ME, Jr PFC, Wagner JA (2008) Comparative inhibitory activity of etoricoxib, celecoxib, and diclofenac on COX-2 versus COX-1 in healthy subjects. J Clin Pharmacol 48:745
Cryer B, Feldman M (1998) Cyclooxygenase-1 and Cyclooxygenase-2 selectivity of widely used nonsteroidal anti-inflammatory drugs. Am J Med 104:413
Pantziarka P, Sukhatme V, Bouche G, Meheus L, Sukhatme VP Repurposing drugs in oncology (ReDO) - diclofenac as an anti-cancer agent, ecancer 10 (610)
Vane JR (1971) Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231(25):232
Ku EC, Wasvary JM, Cash WD (1975) Diclofenac sodium (gp 45840, voltaren), a potent inhibitor of prostaglandin synthetase. Biochem Pharmacol 24:641
Gund P, Shen TY (1977) A model for the prostaglandin synthetase cyclooxygenation site and its inhibition by anti-inflammatory aryl acetic acids. J Med Chem 20:1146
Sallmann AR (1986) The history of diclofenac. Am J Med 80:29
Moser P, Sallmann A, Wiesenberg I (1990) Synthesis and quantitative structure-activity relationships of diclofenac analogues. J Med Chem 33:2358
Beckett AH, Casy AF (1954) Synthetic analgesics – stereochemical considerations. J Pharm Pharmacol 6:986
Castellari C, Ottani S (1997) Two monoclinic forms od diclofenac acid. Acta Cryst C53:794
Kenawi IM (2006) DFT analysis of diclofenac activity and cation type influence on the theoretical parameters of some diclofenac complexes. J Mol Struct 761:151
Rozas I (2007) On the nature of hydrogen bonds: an overview on computational studies and a word about patterns. Phys Chem Chem Phys 9:2782
Bertolasi V, Gilli P, Ferretti V, Gilli G, Vaughan K (1999) Interplay between steric and electronic factors in determining the strength of intramolecular resonance-assisted NH-O hydrogen bond in a series of beta-ketoarylhydrazones. New J Chem 23:1261
Kuo HW, Tsai S, Tiao MM, Liu YC, Lee IM, Yang CY (2010) Analgesic use and the risk for progression of chronic kidney disease. Pharmacoepidemiol Drug Saf 19(7):745
Salvo F, Fourrier-Reglat A, Bazin F, Robinson P, Riera-Guardia N, et al MH (2011) Cardiovascular and gastrointestinal safety of NSAIDs: a systematic review of meta-analyses of randomized clinical trials. Clin Pharmacol Ther 89(6):855
Altman R, Bosch B, Brune K, Patrignani P, Young C (2015) Advances in NSAID development: evolution of diclofenac products using pharmaceutical technology. Drugs 75:859
Kovala-Demertzi D (2000) Transition metal complexes of diclofenac with potentially interesting anti-inflammatory activity. J Inorg Biochem 79:153
Majerz I, Trynda-Lemiesz L (2015) Copper(II) ion as modulator of the conformation of non-steroidal anti-inflammatory drugs. Theoretical insight into the structure. Polyhedron 98:137
Kyropoulou M, Raptopoulou CP, Psycharis V, Psomas G (2013) Ni(II) complexes with non-steroidal anti-inflammatory drug diclofenac: Structure and interaction with DNA and albumins. Polyhedron 61:126
Sahra K, Dinar K, Seridi A, Kardi M (2015) Investigation on the inclusion of diclofenac with beta-cyclodextrin: a molecular modeling approach. Struct Chem 26:61
Jr. IIK, Ovsyannikova LV, Nikulina OI, Belyatskaya AV, Krasnyuk II, Kharitonov YY, Grikh VV, Korol’ LA, Obidchenko YA, Vorob’ev AN (2015) Solubility of diclofenac acid form from solid dispersions. Pharmaceut Chem J 48(11):735
Car R, Parrinello M (1985) Unified approach for molecular-dynamics and density-functional theory. Phys Rev Lett 55:2471
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JAJ, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ Gaussian 09, Revision D.01, Gaussian, Inc., Wallingford CT
http://www.cpmd.org/, CPMD.
Grimme S (2006) Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J Comp Chem 27:1787
Ireta J, Neugebauer J, Scheffler M (2004) On the accuracy of dft for describing hydrogen bonds: Dependence on the bond directionality. J Phys Chem A 108:5692
Vanderbilt D (1990) Soft self-consistent pseudopotentials in a generalized eigenvalue forMalism. Phys Rev B 41:7892
Martyna GJ, Tuckerman ME (1999) A reciprocal space based method for treating long range interactions in ab initio and force-field-based calculations in clusters. J Chem Phys 119:2810
Nose S (1984) A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys 81:511
Hoover WG (1985) Canonical dynamics – equilibrium phase-space distributions. Phys Rev A 31:1695
Bader RFW (1990) Atoms in Molecules, A Quantum Theory, Oxford University Press Oxford
Johnson ER, Keinan S, Mori-Sanchez P, Contreras-Garcia J, Cohen AJ, Yang W (2010) Revealing noncovalent interactions. J Am Chem Soc 132:6498
Contreras-Garcia J, Yang W, Johnson ER (2011) Analysis of hydrogen-bond interaction potentials from the electron density: integration of noncovalent interaction regions. J Phys Chem A 115:12983
Keith TA AIMAll (version 13.05.06, professional), TK Gristmill Software, Overland Park KS, USA
Contreras-Garcia J, Johnson ER, Keinan S, Chaudret R, Piquemal J-P, Beratan DN, Yang W (2011) NCIPLOT: a program for plotting noncovalent interaction regions. J Chem Theory Comput 7:625
Humphrey W, Dalke A, Schulten K (1996) Vmd – visual molecular dynamics. J Mol Graph 14:33
Koch U, Popelier PLA (1995) Characterization of C-H-O hydrogen bonds on the basis of the charge density. J Phys Chem 99:9747
Rowlinson SW, Kiefer JR, Prusakiewicz JJ, Pawlitz JL, Kozak KR, Kalgutkar AS, Stallings WC, Kurumbail RG, Marnett LJ (2003) A novel mechanism of cyclooxygenase-2 inhibition involving interactions with Ser-530 and Tyr-385. J Biol Chem 278:45763
Arunan E, Desiraju GR, Klein RA, Sadlej J, Scheiner S, Alkorta I, Clary DC, Crabtree RH, Dannenberg JJ, Hobza P, Kjaergaard HG, Legon AC, Mennucci B, Nesbitt DJ (2011) Definition of the hydrogen bond (IUPAC Recommendations 2011. Pure Appl Chem 83:1637
Kiefer JR, Pawlitz JL, Moreland KT, Stegeman RA, Hood WF, Gierse JK, Stevens AM, Goodwin DC, Rowlinson SW, Marnett LJ, Stallings WC, Kurumbail RG (2000) Structural insights into the stereochemistry of the cyclooxygenase reaction. Nature 405:97
Llorens O, Perez JJ, Palomer A, Mauleon D (2002) Differential binding mode of diverse cyclooxygenase inhibitors. J Mol Graph Model 20:359
Acknowledgments
We gratefully acknowledge the financial support of the National Science Centre, Poland, grant number DEC-2014/13/B/ST8/04342. Computations were carried out at the Computer Center of University of Bialystok.
Author information
Authors and Affiliations
Corresponding author
Additional information
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kozlowska, M., Rodziewicz, P. & Kaczmarek-Kedziera, A. Structural stability of diclofenac vs. inhibition activity from ab initio molecular dynamics simulations. Comparative study with ibuprofen and ketoprofen. Struct Chem 28, 999–1008 (2017). https://doi.org/10.1007/s11224-016-0893-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-016-0893-8