Abstract
The benefits of physiological biomarkers, knowledge and concepts are well-established in fish and wildlife management as they confer the ability to understand mechanistic processes, identify cause-and-effect relationships, and develop predictive models. Although this approach is gaining momentum in the context of species conservation, the use of physiological biomarkers in exploited marine fish stock management and recovery plans remains relatively rare. Here, we present five essential issues to consider to implement physiological biomarkers in fisheries management: (i) choice of relevant biomarkers that have a well-known mechanistic basis, (ii) identification of species-specific biomarkers reflecting a meaningful timespan for management, (iii) selection of biomarkers compatible with data collection during routine scientific fisheries surveys, (iv) use of biomarkers as early-warning signals and complementary indicators of population-level changes in life history traits and (v) how physiological biomarkers may help to refine long-term population dynamic projections under climate change and management scenarios. Overall, if based on well-established mechanisms linked to individuals’ fitness, a focus on physiological biomarkers should help to better understand the mechanisms behind stock declines, changes in stock characteristics, and thus more efficiently manage marine fisheries and conserve populations. As this approach is transferable among species, locations, and times, the integration of physiological biomarkers in fisheries science has the potential to more broadly enhance assessments and management of fish stocks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding how individuals respond to environmental changes and the mechanisms shaping their life history traits is important for fish and wildlife management (Wikelski and Cooke 2006; Madliger et al. 2016; Bergman et al. 2019). As physiological responses to environmental variability underpin individual life history traits and hence population dynamics (Ward et al. 2016), failing to account for the physiological constraints set by the environment may lead to inaccuracies in our understanding of population dynamics and the management advice derived therefrom (Link et al. 2020). For instance, the decline in reproductive success in birds of prey due to their exposure to dichlorodiphenyltricholoroethane (DDT) led to its ban and the subsequent recovery of these populations (Faroon et al. 2002). Intact glades allow better growth and reproduction (through better assimilation rate) for the Eastern Collared Lizard (Crotaphytus collaris), and thus should be prioritized for conservation purposes (Brewster et al. 2020). Likewise, physiological biomarkers showed that six hours in a salt water pool facilitated post-release survival of Kemp’s ridley sea turtle (Lepidochelys kempii) after transportation (Hunt et al. 2019). Collectively, these (and other studies—see Madliger et al. 2016) studies illustrate the potential of physiological biomarkers, knowledge and concepts to inform the management of species with conservation concerns, an approach that can be adapted to the management of exploited species.
The consideration of physiological processes in recovery plans is highly variable among taxa, with it being somewhat common for reptiles and mammals while relatively rare for exploited marine fish stocks (Mahoney et al. 2018). Most applications of physiological biomarkers in fisheries sciences have focused on the reduction of bycatch mortality arising from commercial or recreational fisheries. For instance, Farrell et al. (2001) demonstrated that placing coho salmon (Oncorhynchus kisutch) in a recovery box decreased their stress levels (measured as plasma cortisol and muscle lactate) and increased their post-release survival. Likewise, Skomal (2007) showed the effects of angling time on multiple physiological factors (blood acid–base status, metabolites and proteins quantities) and provided recommendations to improve the post-release survival of large pelagic fish. Beyond post-capture survival studies where fish have to be released alive, the large number of sampling events carried out by scientific surveys and underpinning many stock assessments and management decisions (Maunder and Piner 2014; Trenkel et al. 2019) opens avenues for the use of multiple physiological biomarkers in fishery science.
Linking physiological biomarkers and fishery science, however, presents difficulties that might stem from the objectives of fishery science (e.g. measuring population productivity) and its need to estimate individual fitness (i.e. the genetic contribution of an individual to the next generation) and population vital rates (e.g. reproductive success with recruitment rate, individual growth rate, and total survival rate). If fisheries data accurately estimate fitness variation and physiological biomarkers are themselves strongly linked to fitness, physiological biomarkers may therefore be perceived as an unnecessary detour to understand population dynamics. Yet, it is well-acknowledge that fisheries data provide imperfect fitness proxies and that typical metrics used in fisheries science such as size at age one, morphometric body condition indices or abundance reveal primarily only rather extreme changes in population ‘state’ (Katsanevakis et al. 2012; Breton et al. 2013; Maunder and Piner 2014). For instance, even when environmental conditions become suboptimal and the constraints of maintaining internal conditions within physiological tolerance bounds increases (Pérez-Ruzafa et al. 2018), mechanisms such as compensatory or catch-up growth (Ali et al. 2003) and/or trade-offs between life history traits (growth vs survival; Sogard 1997) may lead to an apparent stasis in population phenotypic characteristics (size-at-age, body condition). These mechanisms, however, mask fundamental changes in population state (e.g. size-dependent mortality) and density dependence effects that might only become evident later on, delaying the implementation of measures to improve stock management and conservation (Lennox et al. 2018). Consequently, managers respond to declining populations by changing management measures without necessarily understanding the mechanisms behind population declines, potentially leading to inefficient results (Mahoney et al. 2018). To this end, physiological biomarkers that react quickly to environmental changes and management actions could be used to define thresholds or reference points for fisheries science, and may provide much-needed information, for example, on how the trajectory of a population may be linked to reduced growth and survival.
A variety of physiological biomarkers, each potentially equally important, affect or provide information on body condition and life history traits from an evolutionary ecology point of view (Kotiaho 2002; Tomkins et al. 2004; Mangel and Munch 2005). Yet, fishery scientists often restrict the notion of an individuals’ body condition to the amount of stored energy (e.g. Clancey and Byers 2014). Individual body condition and life history trait variability have been related to individuals’ energy reserves (from energy density to fatty acids profiles, Parrish 2009) but also to metabolism (Rowe and Houle 1996), parasite load (Timi and Poulin 2003), immune response (Zuk and Stoehr 2002), ability to sustain physical efforts (Johnstone et al. 2017), nutritional stress (Hulbert et al. 2014), oxidative stress levels (Costantini 2008), and overall stress levels (Korte et al. 2005). Due to the multitude of pathways affecting physiology and life history traits (Ricklefs and Wikelski 2002), the use of multiple biomarkers carefully selected based on well-known mechanisms can enable the characterization of individual overall health status (Cooke and O’Connor 2010). Indeed, even if the application of physiological biomarkers to fisheries questions only generate correlative findings, changes in physiological biomarkers can provide rapid information regarding individual and population responses to environmental conditions and serve as early warning signals (e.g. cortisol, lipid content, metabolic rate, parasite load; Coristine et al. 2014). Hence, it is essential to focus on several physiological biomarkers with clear mechanistic bases related to a given degree of impairment of life history traits to better understand direct links between environmental conditions, physiological biomarkers, and individual life history traits to predict population-level outcomes. Therefore, bridging the gap between physiology and fishery science has the potential to improve our understanding of exploited species’ population dynamics, enhance our ability to predict population responses to environmental changes, and hence support management decisions (Martino and Houde 2010; Madliger et al. 2016; Nash et al. 2021).
The contribution of physiological research to fisheries sciences has previously been highlighted (see Young et al. 2006; Metcalfe et al. 2012; Horodysky et al. 2015; McKenzie et al. 2016). However, these papers often focus on a single technology or on the way to translate individual-level physiological biomarkers into population-level processes. Here, our goal is to provide a framework to guide the choice of physiological biomarkers with the greatest potential to be relevant for aquatic fisheries management (Colin et al. 2016). These relevant biomarkers (i) have a clear mechanistic basis linked to life history traits variability, (ii) are unaffected by the capture and sampling processes, (iii) can be implemented within the routine operations of scientific fisheries surveys, (iv) can be used as early warning signals of population changes in life history traits, and (v) can improve long-term projections of population demography as well as associated management strategies. Our aim is not to provide a summary of all possible applications of physiological biomarkers in fisheries science, but rather to illustrate the diverse ways in which these biomarkers have the potential to enhance fisheries management.
Physiological biomarkers for fisheries science
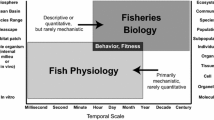
Before being translated at population scales (relevant for fisheries management), two key properties are essential for physiological biomarkers: (i) a clear mechanistic basis and known effects on individual fitness (i.e. experimental and theoretical cause-and-effect relationships) and (ii) being easily collected in the field and accurately measurable. The physiological biomarkers presented below are thus discussed in terms of their significance for measuring individual fitness and their implementation during routine fisheries surveys.
Mechanistic basis of physiological biomarkers
Here we provide several examples of physiological biomarkers, from whole-organisms to gene expression markers. These examples are by no means exhaustive but reflect the ones that are known to date to be linked to life history traits and hence have the greatest potential to be relevant for fisheries management (i.e. metabolism, stress responses, energy reserves quantity and quality, immune responses, oxidative stress responses, and -omics; see also Table 1).
Metabolism is a key process converting nutrients into energy usable for different functions and therefore determines individual energy acquisition, transformation and allocation to different life history traits (e.g. growth, reproduction; Newsholme and Start 1973). In species for which individuals can be released after capture following sampling, estimating the metabolic rate indirectly (i.e. typically measured as the rate of oxygen uptake or the heart rate) in their natural habitat is possible with boat or shore based respirometry (Chabot et al. 2016) and/or electronic tags (Metcalfe et al. 2016). These methods are useful for understanding the behaviour and energy allocation of wild fishes as a function of environmental conditions (Chabot et al. 2016). Fish metabolism is also affected by hormones such as glucocorticoids that will rapidly rise in response to stress factors (within 3–5 min) and have provided some of the most successful set of animal stress biomarkers (Denver et al. 2009). Monitoring basal corticosteroid level or the increase in corticosteroids after a standardised stressful event in various tissues such as muscle, blood, eggs, mucus or scales have been found to be valuable biomarkers of short-term and chronic stress (Baker et al. 2013; Sadoul and Vijayan 2016). Assessing chronic stress is particularly interesting due to its detrimental consequences for the metabolism, the immune system (Korte et al. 2005; Sadoul and Vijayan 2016), growth (Gregory and Wood 1999) and for reproduction (Faught and Vijayan 2018). Alternatively, the metabolic activity over a timespan of 2–3 months and feeding intensity over the last 48 h in fish can be estimated using pancreatic and intestinal enzymes (e.g. trypsin and alkaline phosphatase) that facilitate the hydrolysis and absorption of nutrients, respectively (Cahu and Zambonino-Infante 1994; Lemieux et al. 1999). For these reasons, digestive enzymes have been widely used as biomarkers of fish physiological functions related to growth and survival abilities (Ueberschär 1995; Zambonino-Infante et al. 2008; Zambonino-Infante and Cahu 2010).
The amount of energy reserves stored in a whole individual can be directly measured (e.g. bomb calorimetry) or approximated based on the total amount of lipid extracted from key energy storing organs (e.g. gut, muscle, liver). These metrics have proven useful to assess the energetic status of individuals and to address question related to life history strategies, ecology and fish stock management (Lloret et al. 2014). These approaches are, however, limited in a number of ways as only a fraction of the energy measured in the entire body may be allocated to the different functions (Sargent et al. 1999; Tocher 2003) and energy reserves may be stored in different forms (glycogen vs lipids) during development (Josrgensen et al. 1997), making it difficult to quantify the actual amount of energy reserves available during growth (Jobling and Johansen 1999). In addition to differences in the amount of energy reserves, differences in their quality may be important as for instance the availability of essential lipids that support crucial physiological functions can influence directly the development and health of fish. Especially, dietary n-3 polyunsatured fatty acids (omega-3) composition can affect growth, reproduction, behaviour, vision, stress resistance, disease resistance and immune response (Kiron et al. 1995; Sargent et al. 1999; Montero et al. 2003). Therefore, monitoring individual fatty acids composition could be more accurate to estimate individual health.
As immune responses are costly to mount, the optimal response to a pathogen will depend on an individuals’ current condition and infection status (Viney et al. 2005). A simple way to evaluate immune response is to quantify the lysozyme activity (Lie et al. 1989) or the neutralizing antibodies (e.g. using a cell-based assay; Tort et al. 1996; Boshra et al. 2006). The immune system is a network that protects the organism against diseases and pathogens. Thus, measures of individual immune responses are key in understanding variation in health status and hence the consequences on life-history traits (Sheldon and Verhulst 1996; Šimková et al. 2008; Dupuy et al. 2015). Oxygen is essential for many metabolic processes but oxygen metabolism also leads to the inevitable production of reactive oxygen species (ROS; Sies 1997). ROS act as signalling molecules to regulate biological and physiological processes with roles in apoptosis and the induction of defence genes transcription. However, when the amount of ROS overcomes an organisms’ anti-oxidant capacities, for example during rapid individual growth or lasting exposure to stress factors, ROS will damage lipids, proteins and/or DNA (oxidative stress; Yu 1994; Costantini 2008). Such unbalanced situations will have negative consequences on individual growth, reproduction and body maintenance (Birnie-Gauvin et al. 2017). Oxidative stress levels can be measured using biomarkers of damage (e.g. lipid peroxidation, protein carbonylation, DNA-damage), and endogenous or exogenous anti-oxidant defences (e.g. superoxide dismutase, catalase, carotenoids; Beaulieu and Costantini 2014).
Among molecular level biomarkers, metabolomic, proteomic, transcriptomic and genomic tools are the most recently developed ones (Madliger et al. 2018; Bernos et al. 2020; Le Luyer et al. 2021). This suite of ‘-omics tools’ can pinpoint key metabolites, microbiome dysbiosis, proteins and genes involved in individual response to changes in the environment. For example, yolk proteins have been used as biomarkers of endocrine disruptions that impair fish reproduction (Kime et al. 1999). Likewise, plasma proteomics have allowed the detection of a network of protein changes associated with immunological pathways in stressed fish and may be biomarkers of chronic stress (Raposo De Magalhães et al. 2020). These ‘-omics tools’ therefore facilitate the investigation of individual response to environmental changes, and the establishment of causal links between molecular and physiological/organismal variation. The main constrains remain the need to quickly freeze and keep the sample frozen at − 80 °C in order to ensure proteins or RNA integrity and relatively high analysis cost that limit sample size.
The effects of specific environmental conditions on many of these physiological biomarkers are well established under laboratory conditions (Vinagre et al. 2012; Vanderplancke et al. 2014; Birnie-Gauvin et al. 2017). However, different environmental factors (e.g. increasing water temperature and decreasing oxygen concentration) can have additive, synergic or antagonistic effects on individual physiology and physiology–fitness relationships (Wernberg et al. 2012; Kimball et al. 2012; Côté et al. 2016). Moreover, individual behaviours such as dispersal or foraging decisions can directly dampen the effects of adverse environmental conditions and trigger other physiological responses (e.g. climate change shifted distribution areas may have higher predator densities and thus lead to more fish chronic stress; Tuomainen and Candolin 2011). Therefore, in addition to the development of a roadmap for successful use of physiological biomarkers in fisheries management (see below), a concerted effort combining in-situ data as well as controlled laboratory studies is necessary to calibrate in-situ measures. Indeed, we need to derive more knowledge on potential climate change-related drivers impacts on several vital rates from laboratory and field studies (e.g. with ecotoxicological measures on lethal concentration of a substance in water causing a death of 50% of the tested population, LC50) to build robust models for climate impacts projections. These models could adequately assess physiological constraints and mechanisms underpinning individuals’ health and vigour for a large number of natural populations and provide science-based advice for sustainable management of aquatic resources (Peck et al. 2013; Catalán et al. 2019).
Enable scientific survey samples to be physiology-compatible
The collection of samples required to measure these physiological biomarkers must be easily implemented within the routine operations of scientific fisheries surveys (see the “Fishery survey compatibility” step in Fig. 1). The most common constraints for onboard sampling are time, space, and sample storage facilities. Consequently, physiological measurements less impacted by tissue degradation and/or requiring a small quantity of tissues and equipment should be prioritised (see Table 2) over biomarkers based on whole organism performance such as classical respiratory experiments which would require equipment rarely available onboard (even though some recent innovations propose promising practical and reliable field respirometry methods; Mochnacz et al. 2017) or on shore but outside of traditional academic laboratory environment. There have also been innovations in sampling that allow use of small, non-lethal tissue biopsies (e.g., gill tissue for transcriptomics work, Jeffries et al. 2021; blood for use in say cortisol assays, Lawrence et al. 2020) such that data can be collected from rare or threatened species that are released alive.
The major issue with samples collected during fisheries surveys is the impact of acute stress response associated with the capture process on many physiological biomarkers. Irrespective of the fishing gear, physiological biomarkers strongly affected by these processes must be avoided because the acute (and short-term) stress responses to fishing are bound to swamp the longer-term baseline levels of these biomarkers. For instance, hormonal concentrations in plasma or leukocyte counts can change rapidly in response to fish exhaustion and stress (Dhabhar 2002; Walker et al. 2005) while the lytic activity of plasma leukocytes or the concentration of scale cortisol are largely unaffected by the acute stress responses of fish and hence largely unaffected by capture (Sadoul and Geffroy 2019). It might also be possible to statistically account for these effects by using an estimate of ‘capture stress’ (see Lawrence et al. 2018 for example) but this would be done based on the assumption that the response of these biomarkers was consistent across individuals, an assumption that might be impossible to experimentally assess in many species. Thus, biomarkers that are not influenced by fishing acute stress such as scale cortisol or lipid stores should be favored. Moreover, onboard workload often implies delays in sample collection after capture that induce a rapid degradation of fish tissues. The sensitivity of each physiological biomarker to this degradation must be evaluated and it is critical to identify the maximum time during which a sample is amenable to sampling for specific biomarkers and environmental conditions. Recent studies that focus on genomics or transcriptomics tend to use RNA stabilization solution as a preservative and although refrigeration is preferred, it is not essential. Every biomarker, test, and tissue type is different in terms of how it should be preserved and physiological biomarkers with the least demanding requirements should again be favored. For instance, samples collected to investigate oxidative stress levels only need to be rapidly placed in − 20 °C freezers and analysed within 4 months (Beaulieu and Costantini 2014), the activity of digestive enzymes can relatively easily be preserved using non-toxic reagents (Cahu and Zambonino-Infante 1994), and the use of lytic activity of plasma leucocytes requires only storing plasma samples at − 20 °C. If no processing facilities are available onboard, freezing the whole fish is the only solution to bring back exploitable samples to the laboratory. In this case, one must select a subset of biomarkers that are the least affected by freezing and thawing processes.
Steps to follow in the selection of optimal physiological biomarkers
After addressing the issues of reliability and feasibility, the different responses of biomarkers to stress factors points to the advantages of employing a suite of biomarkers. Obviously, the success of such an approach would depend on the choice of this suite of biomarkers. Following Cury and Christensen (2005) and Rice and Rochet (2005) approach, we focus on two decisive steps to choose physiological biomarkers intended to support management decision-making: (i) the identification of biomarkers adapted to the target species, with an appropriate temporal integration (i.e. the timespan over which the biomarker reflects a given physiological process), informative for management, and with both reasonable financial cost and time investment, and (ii) the selection of the most relevant suite of biomarkers for providing information on individual and population changes for the considered species.
Identification of species-specific physiological biomarkers with temporal integration relevant for management
Wild fish populations may have different ecological features, inhabiting a single or a diversity of habitats, and displaying a variety of behaviours. Therefore, the choice of physiological biomarkers has to be tailored to each species, accounting for characteristics such as size/life history stage (that constrains the amount of tissue that can be collected), habitat (i.e. demersal or pelagic) or functional traits (e.g. dietary preferences, dispersal abilities, longevity), and even population and sex. For instance, the continuous swimming activity of pelagic fish makes them highly sensitive to oxygen availability while strictly demersal fish will be less sensitive to this parameter (Clark et al. 2013; Brown and Thatje 2014). Likewise, life at the poles has led to a wide range of physiological adaptations in fish. For instance, Antarctic icefish (Notothenioidei sp.) such as the loss of red blood cells and haemoglobin as a result of living in oxygen-rich waters (Cheng and Detrich 2007). Such key ecological differences will make some biomarkers more informative than others depending on the studied species. Furthermore, fish anatomic properties can limit the sampling of some physiological biomarkers. For instance, fish species without scales or with small scales that are difficult to sample (e.g. flatfish) or often renewed (small pelagics) are not appropriate to study cortisol content in scales. Moreover, small-sized species may be unsuitable to the collection of some biomarkers because of the limited amount of tissue that can be sampled (e.g. about 50–75 mg of fish scale is needed to quantify cortisol concentrations, Sadoul and Geffroy 2019). In such cases, it is possible to pool individuals to obtain enough material at the expense of information at the individual level and the risk of averaging fish with different physiological states. Another key point is the deployment of physiological biomarkers for different life stages. For example, population productivity depends on adults (e.g. maternal effect) and the survival of their offspring during early life stages (Brosset et al. 2020). Therefore, it might be possible to improve recruitment forecasts by studying both larval, juvenile, and adult life stages to get a more holistic knowledge and strengthen our ability to use them for population management. These species and life stage-dependent aspects must be considered from the beginning when selecting a physiological framework as they make the choice and the feasibility of optimal biomarkers highly species-specific (step “Biomarker selection” in Fig. 1).
Another key aspect of physiological biomarkers selection is the temporal scale of the processes they reflect. This temporal integration period, referred to as timespan, depends on the way physiological processes are regulated and hence the speed at which a biomarker responds to environmental variations (e.g. stress hormones: quick increase and then a steady decline once the stress factor has disappeared; conversely heat shock proteins take much longer to build up and to get back to their baseline, Madliger et al. 2018). Depending on the tissue sampled (e.g. blood, muscle, eggs, or organ biopsy, see Madliger et al. 2018), the reference time period of a given physiological biomarker may vary. For instance, information for different reference periods can be obtained when measuring cortisol levels in blood (snapshot at the time of sampling), in eggs (maternal stress level over previous weeks) or in scales (stress level integrated over an individual’s lifespan; Aerts et al. 2015). Similarly, measurements of metabolism can be based on enzyme activity with a timespan of ca. 2–4 weeks or fatty acids contents that reflect longer term diet and metabolism (Parrish 2009). Depending on the question asked, short-term physiological biomarkers may be more useful for investigating acute stress at a particular life-stage or geographic location as they allow identifying a specific environmental pressure, while long-term biomarkers will most likely describe broader life history constraints.
Once a set of suitable physiological biomarkers for a given species has been identified, it is critical to ensure that measurements are accurate. Multiple samples should be collected and technical replicates for a subset of individuals should be measured to quantify sampling variation within individuals and hence the precision of a given physiological biomarker. As tissues can rapidly deteriorate, repeatability has also to be quantified according to the time elapsed between an individual’s death and sample collection. Some biomarkers allow the direct quantification of tissue degradation (e.g. the appearance of free fatty acids, Couturier et al. 2020) but in many instances such degradation will result in a decline in the magnitude of the measurement (e.g. blubber cortisol, Trana et al. 2015). Conversely, physiological biomarkers must show sufficient inter-individual variability to enable us to characterise differences in health state.
Selection of the most powerful physiological biomarkers
The selected physiological biomarkers must add novel insights to the classical measurements carried out during fisheries surveys. Indeed, the relationships between physiological and size-at-age or body condition measures are not necessarily straightforward and physiological biomarkers may highlight constraints and trade-offs that cannot be revealed by morphometric measurements alone (Kimball et al. 2012). Consequently, physiological biomarkers could be assessed in a correlative way with the different life history traits (or their proxies) routinely measured during fisheries surveys (e.g. size-at-age, body condition, and reproductive status) to identify the shape and strength of these relationships.
The robustness of inferences drawn from physiological biomarkers is enhanced by the integration of complementary biomarkers, ideally across multiple levels of biological organization, that encompass several physiological mechanisms and underpin individual fitness (Trenkel et al. 2007; Hook et al. 2014). Physiological biomarkers are, however, often considered separately while the constraints resulting from changes in health and acting on life history traits are clearly multifaceted (e.g. on rapid growth; Metcalfe and Monaghan 2003). Combining multiple physiological biomarkers is therefore necessary to understand the interaction between life history traits and the overall consequences of environmental changes. Thus, the use of several biomarkers would likely decrease the probability of a false positive due to a spurious result in a single biomarker (Hook et al. 2014; Madliger and Love 2015). For instance, previous work has shown that increasing digestive enzyme activity is related to faster fish growth which itself is negatively related to oxidative stress (Metcalfe and Monaghan 2003). Similarly, individuals having higher chronic stress levels may have lower growth rates and body condition resulting from the increasing energetic demand of mounting the stress responses (Abdel-Tawwab et al. 2019). Thus, the choice of the number of biomarkers has to be done carefully and should rely on knowledge of physiological links between them. This can be done through the implementation of multivariate analyses (e.g. principal component analysis, correspondence analysis or factor analysis, Path-Analysis) to estimate the covariation between physiological biomarkers, and identify the most powerful biomarkers (Greenstreet et al. 2012). Multivariate analyses can also enable us to extract a few compound variables describing individual overall health state which might in turn impact life history traits (i.e. multivariate eigenvectors may readily reflect individuals’ health state combining for example low parasite load, good nutritional state and low chronic stress). As for each physiological biomarker, the eigenvectors resulting from the multivariate analyses could be correlated with different life history traits (or their proxies) to determine whether they outperform separate physiological biomarkers (i.e. to highlight undetected mechanisms). This approach might be useful in cases of snapshot information but such compound variables are not directly comparable overtime if they are defined based on correlations/covariances between variables in separate years. Other methods to combine indicators are proposed in Rice and Rochet (2005) and could also be considered. A flowchart summarizing the main steps of the biomarker selection and validation is provided in Fig. 1. Overall, researchers and managers must weigh costs and benefits when deciding whether a given physiological biomarker is applicable in their system.
Using physiological biomarkers in fisheries management
Physiological biomarkers have been largely used in ecotoxicology and especially in the context of chemical contaminant exposure, noise pollution, or fish post-release mortality (Adams et al. 1992; Scott and Sloman 2004; Skomal 2007; Gagliano and McCormick 2009; Sierra-Flores et al. 2015). However, little attention has been paid to their potential application in fisheries management. The main hurdle to overcome is the scaling up from individual physiology to population processes and the integration of these biomarkers in the decision-making process (Hamilton et al. 2016; Bergman et al. 2019). Because of their mechanistic basis, sustained and substantial changes in physiological biomarkers are bound to influence individual life history traits in a linear or non-linear way (i.e. continuous change vs thresholds specific to each biomarker). Thus, physiological biomarkers can indicate changes in population dynamics that would have been missed by the mere monitoring of morphometric changes at age and reproductive status and become physiological biomarkers of current and future population changes (Monaco and Helmuth, 2011). Here, we discuss a number of case studies that highlight the importance of considering physiological biomarkers to understand population demography and to forecast long-term population changes under global change and fishing scenarios. Koenigstein et al. (2018) showed that understanding the physiological basis of the mortality of early life stages to temperature and acidification allowed to investigate warming and acidification impacts on Barents Sea cod recruitment to fisheries and project future stock dynamics. Thus, severe reductions in average age-0 recruitment success and population decline of Barents Sea cod are projected under uncompensated warming and acidification towards the middle of this century (Koenigstein et al. 2018). Still focusing on early life stages survival, Moyano et al. (2020) found that increasing temperature led to a decline in the cardiac performance and growth rate of herring larvae which were associated with declines in population productivity and proposed as indicators for population resilience. We now outline how physiological biomarkers could inform fisheries management and conservation and help understanding how environmental and fishing changes influence exploited marine populations (Fig. 2).
Theoretical framework to make physiological biomarkers useful for fisheries management. With regard to the shape of the curves, they may be smooth for some species, but could equally well be a stepwise relationship in which a species can compensate for disturbance up to a point and then abruptly transitions into a new physiological status
Identifying ‘early warning’ physiological biomarkers
In fisheries science, the indicator approach aims at adjusting management strategies based on indicators characterizing the impacts of environmental variations and human practices on exploited populations (Jennings 2005; Rice and Rochet 2005; Rochet et al. 2005). The use of this approach is growing worldwide as it provides a robust, simple, and pragmatic assessment tool for many freshwater and marine stocks (Beauchard et al. 2017; Thompson et al. 2020). However, to date few studies have sought to integrate physiology into this approach (Adams and Ham 2011; Jeffrey et al. 2015) while there is an increasing number of physiological studies identifying sublethal and lethal effects of climate change and fishing (Bozinovic and Pörtner 2015). Moreover, physiological states do not necessarily vary linearly, requiring the identification of thresholds in organismal- and then population-level performances according to the environmental pressure(s) experienced (i.e. thresholds beyond which growth is impaired or survival and/or reproduction is diminished, Ricklefs and Wikelski 2002).
Physiological biomarkers complementary to current fishery indicators could be identified based on the relationship between physiological biomarkers and life-history traits (Table 1). For instance, antioxidant defences are typically associated with higher fertility and survival rate, while oxidative stress is associated with a decrease in reproduction and growth, and hence the current and future fitness of individuals (Costantini 2008). Likewise, elevated glucocorticoid(s) levels have been associated with reduced fish survival, poor gamete quality and poor reproductive success (e.g. cortisol; Baker et al. 2013). The physiological biomarkers used to study fish post-release survival (Horodysky et al. 2015) might also be useful to estimate discard mortality and hence be used to improve estimates of unaccounted fishing mortality (except for European stocks for which the landing obligation applies). Investigating such performance curves (e.g. variation in growth, reproductive success and survival along environmental gradient that define an organism's critical limits) is particularly relevant to understand how fish cope with environmental change and fishing, and ultimately examine population dynamics and their change. Thus, individual overall stress levels and physiological responses can help us to understand population level tolerance to natural stress factors and bring further our conservation and management efforts to mitigate anthropogenic disturbances (Sopinka et al. 2016; Birnie-Gauvin et al. 2017). Usually, biomarkers are measured in a sub-sample of the population under investigation. The mean response of the individuals in the sample is then used to estimate the expected response of the whole population. In formulating new biomarker approaches, it is also important to move beyond measuring mean responses or the mean effects that ensue. The proportion of individuals in the population below or above physiological biomarker thresholds underlining adverse effects of the environment should thus be considered. Indeed, the success of natural populations and communities are determined by the range of attributes of their individuals, and the determination of the relative proportions of biomarker levels in regard to thresholds can be used as a first insight into population-level consequences of environmental changes and potentially yield insight on their adaptive capacity (Depledge et al. 1993).
Physiological biomarkers may also be used to reveal yet undetected changes when for example there are signs of recovery at higher levels of biological organization (populations, communities) while biochemical and physiological systems remain impaired (and vice versa). As physiological mechanisms cannot fully compensate over an extended period of time, the deleterious effects of environmental changes on fish life history traits and fitness might appear with a delay (‘carry over effects’, e.g. compensatory growth has been observed to cause long-lasting alterations to basal metabolic rate; Killen 2014). Moreover, directional and disruptive selection caused by anthropogenic or environmental factors might lead to permanent changes in physiological processes if their levels and regulation are heritable. Beyond highlighting underlying mechanisms responsible for fish phenotype changes, most physiological biomarkers rapidly respond to the multiple environmental and anthropogenic changes and pressures experienced by individuals and can therefore provide early warning signals of environmental deteriorations, relapses and recovery dynamics. Highlighting such rapid dynamics could serve exploited stocks for which management rely only on indicator-based assessment methods (e.g. abundance, mortality rate, length and age compositions; Cooke and Suski 2008; Cotter et al. 2009). Another strength of physiological biomarkers is their ability to identify the mechanism(s) impacted by multiple pressures (Killen et al. 2013). Indeed, the relationships between environmental factors and physiological biomarkers are likely to be stronger than those obtained with common morphometric indicators such as growth parameters derived from length-at-age or indices of body condition. Multiple physiological biomarkers could routinely inform management on the drivers of observed population dynamics, act as early warning signals, and thus contribute to proactive fisheries management.
Finally, physiological biomarkers and their benefits listed above may complement the efforts to protect the marine ecosystem and biodiversity upon which our health and marine-related economic and social activities depend (e.g. the 11 descriptors of “Good Environmental Status” in the European Marine Strategy Framework Directive). By providing new and early-warning signals about mechanisms shaping individual fitness and population dynamics, they could facilitate integrated stock and ecosystem health status assessments. The use of scale cortisol measures as chronic stress biomarker for keystone fish species, for example, could be a powerful way to monitor environmental change and anthropogenic disturbances altering ecological functioning and thus health status of communities and ecosystems.
Physiological biomarkers and population modelling
Physiological biomarkers may improve projections of both environmental and fishing effects on individual fitness and hence population dynamics. Understanding these physiological responses and the associated variability of life history traits combined with projections of future environmental conditions might be invaluable to predict the vulnerability of populations to climate change and thereby the sustainability of the fishery (by direct effects on the physiology of individuals and through trophic cascades; Pérez-Ruzafa et al. 2018; Little et al. 2020). Indeed, the effectiveness of management plans depends on reliable predictions of life history traits’ variability and species’ range shifts outside the range of available (historical) data. Thus, management plans failing to account for past and future changes in life-history traits and range shifts will likely misestimate long-term yields or miss rebuilding targets. For instance, changes in the spatial distribution of fish stocks may greatly affect the uncertainty of biomass projections if the processes driving life history changes are not quantified accurately (Mangel and Levin 2005; Monaco and Helmuth 2011).
One way to inform extrapolation in model projections is to model explicitly the link between environmental variation, physiological processes, and fitness over the entire life cycle and broad geographic ranges in a population context (Baskett 2012; Horodysky et al. 2015). In recent years, there has been renewed interest in physiologically-based life cycle models such as the dynamic energy budget theory (DEB) or biphasic models to assess individual-level effects of lethal but also sublethal stressors (Wilson et al. 2018; Watson et al. 2020). These models allow an understanding of the trade-offs between growth, survival and reproduction at individual level and provide a means to assess the range of interacting drivers (Huebert and Peck 2014; Teal et al. 2018). For instance, several studies have shown that such models can accurately predict growth and reproductive success patterns (Baas et al. 2018). For example, DEB models allow the study of changes in energy pathways resulting from various stress factors (e.g. food availability, water temperature, hypoxia or pollutants). However, because these models specify the response of an individual, tools such as individual-based model (IBM) are needed to extrapolate their effects to population levels. The DEB-IBM framework is designed to explore properties of both individual life‐history traits and population dynamics, their interaction with environmental variables such as temperature or food quantity, tests of DEB theory in a population context, and to advance individual‐based modelling by basing the representation of individuals on well‐tested physiological principles (Bueno-Pardo et al. 2020). Such approaches can thus provide a consistent framework for modeling species physiology and combined with projections of future climate scenarios, predict future population dynamics.
In addition to informing mechanistic forecasting approaches, tuning life-history trait parameters according to individual response to environmental changes in population dynamics models can allow the evaluation of their impacts on management decisions (Cooke et al. 2013). Thus, rather than assuming invariant life history traits (e.g. constant growth or natural mortality), length- or age-based population projection models could incorporate expected changes in life history traits, depending on the climate change and/or fisheries scenarios being considered (e.g. different stress levels affect growth rates, McKenzie et al. 2016; Little et al. 2020). As population dynamics models often use annual estimates of body weight and natural mortality rates at length or age, the link between life history traits and environmental variability (through physiological functions) could be used to adjust size- or age-dependent life history trait values. For example, including these traits in Von Bertalanffy models (based on a bioenergetic expression of fish growth: Von Bertalanffy 1957) may significantly improve simulations for either length or weight data. For spatially explicit models, integrating the spatio-temporal dynamics of the fish–environment interaction (e.g. physiological tolerance curves to different environmental parameters) can improve current estimates and future predictions of stock distribution and abundance (Horodysky et al. 2015). Other examples combine metabolic scope theory with more detailed quantitative models to assess how bioenergetic effects on life history trade-offs might affect populations (e.g. natural mortality; Jørgensen and Holt 2013). Understanding physiological state at early life stages may also be particularly useful for formulating stock–recruit relationships. Indeed, the variation in the number of recruits for a given stock size is one of the main drivers of population dynamics. Accounting for the relationship between environmental conditions and individuals’ physiological characteristics during early life stages could improve the accuracy of survival and growth estimates.
By establishing the mechanistic links between environmental conditions and physiological biomarkers, physiological conditions can thus be used to project changes in vital rates in the long term, for example to evaluate the impact of climate change scenarios. These research topics are promising as they could directly estimate the effect of incorporating physiological knowledge within projection models.
Examples and challenges of including physiological biomarkers in fisheries management
Only a few studies (mainly on amphidromous fish species) showed a practical application of their analyses of physiological biomarkers in management plans. Patterson et al. (2016) used quantitative models relying on physiology measurements such as metabolism and swimming ability under different temperature scenarios to understand the high in-river mortalities of Fraser River sockeye salmon due to water temperature changes. As high mortality events were associated with high migration temperatures, Patterson et al. (2016) forecast how water temperature affects Pacific salmon population in-river mortality each year. This study led to changes in the management model used by fisheries managers for this stock to determine catch levels to achieve the desired level of spawning escapements taking into account predicted losses associated with adverse environmental conditions. Another example refers to the collaboration of physiologists and fisheries scientists to estimate post-release mortality. Farrell et al. (2001) used physiological response variables (such as cortisol and lactate concentration) to show that Coho salmon captured as by-catch in commercial nets placed in a Fraser recovery box for 1–2 h helped their physiological recovery and increased post-release survival (Farrell et al. 2001). Due to the high number of by-catch, this was assumed to facilitate recovery of Coho salmon and to decrease population-level fishing mortality, leading to regulations requiring gill net boats to have recovery boxes to minimize Coho salmon post-capture delayed mortality.
Other studies highlighted the importance of considering physiological biomarkers to improve management outcomes, and although they are not yet operational, these results would deserve attention from managers. For instance, Koenigstein et al. (2018) showed that the severe reductions in average age-0 recruitment success of Barents Sea cod (see above) could be mitigated by management measures sustaining the cod stock at the high spawning stock biomass of recent years. Moyano et al. (2020) recommended to adjust various stock productivity projections according to temperature (linked to cardiac performance of herring larvae, see above) in future management scenarios. Fahd et al. (2021) used contaminant levels to estimate survival rates of adult polar cod, their negative outcome at the population-level as well as their cascading effects on the food web in a risk assessment model investigating various oil spill scenarios. In a more general way, Beaulieu and Costantini (2014) found that oxidative stress status was related to individual fitness and population growth, advocating for a larger use of this type of biomarker as indicators in wildlife management. Similarly, Cheung et al. (2011) combined fish sensitivity to oxygen concentration and ocean acidification to point out that climate change may lower the growth rate of exploited marine fish population and hence reduce catch potential by up to 30% in 2050. These last examples rely on solid knowledge of physiological mechanisms and clearly show how cause–effect approaches can help us to devise more effective management strategies to mitigate the effects of future environmental conditions on abundance and productivity of marine species. In Fig. 3, we represented how various actors could use the pathway proposed in Fig. 1. Using the example of a small pelagic fish population showing biomass and recruitment decline over time, we illustrated the compatibility of physiological biomarkers with fisheries surveys (e.g. we removed basal corticosteroid level due to its high sensibility to capture and metabolic rate which is difficult to measure onboard), the suitable biomarker selection regarding the species and time-scale of interest (e.g. we removed liver and blood biomarkers due to low quantity of energy stored in the liver and short-time scale of the processes evaluated with ROS in blood, respectively) and the potential management actions following changes in physiology (e.g. changes in growth, mortality parameters and recruitment scenario in stock assessment models, Fig. 3).
Selection process for potential management actions given changes in physiology of an exploited population, referring to the pathway presented in Fig. 1. For this example, we retained a theoretical small pelagic fish (SPF) population showing biomass and recruitment decline over time and explored the possibility to use 13 different biomarkers. SAM stock assessment model
A critical drawback for the application of physiological biomarkers in fisheries conservation and management is the lack of long-term monitoring data. Some historical samples stored by the fishing industry or research institutes might be suitable to retrieve information about the levels of some physiological biomarkers (e.g. amount of fatty acids for small pelagic, scales archived after use for ageing might be used to quantify cortisol levels). However, in many cases measuring physiological biomarkers and their potential threshold levels will require first the application of space for time substitutions (and the ensuing theoretical and practical hurdles that such replacement induces; Pickett 1989; Lester et al. 2014). By using a large spatial gradient of existing environmental conditions, it might be possible to obtain a first estimate of the response of physiological biomarkers to variations in environmental factors. Consequently, one of the most important challenges is to gather field information to determine the relationship with fitness proxies for each stock of interest.
Several challenges are associated with the use of physiological biomarkers. Even if the combination of physiological responses to environmental conditions is important for fisheries, considering species’ capacity to compensate for global change is necessary to determine how vulnerable they might be to present and future environmental changes. Indeed, natural populations may show evolutionary responses to climate change (Thomas et al. 2001) and/or phenotypic plasticity. Such effects are addressed in studies looking at fisheries-induced evolution, which can occur if individual fish within a population vary in their vulnerability to capture through differences in traits such as in size, physiology or behaviour, if these differences are heritable (Diaz Pauli et al. 2015). However, most fisheries-induced evolution studies rely on approaches limited to data describing maturity reaction norms or growth curves (Ernande et al. 2004; Nusslé et al. 2009). Differences in physiological performance constraining growth or survival would then not only reflect changes in the environment but also phenotypic plasticity and/or adaptive changes in response to environmental variations. Moreover, individual phenotypic diversity needs to be considered at the ecosystem scale as maintaining or increasing inter-individual trait diversity may improve population resilience to environmental perturbations and hence have positive effects on communities and ecosystems (Ward et al. 2016). Thus, practitioners need to account for ecosystem processes at evolutionary time scales to manage fisheries resources that would be bridged in the long run (Laugen et al. 2014).
Conclusion
Physiological biomarkers have great potential to inform the management of marine fish stocks if applied according to the following guidelines: (a) consider only physiological biomarkers with a clear mechanistic basis and ecological relevance regarding individual fitness; (b) adjust sampling to survey logistic constraints (fast sampling with small and easily storable samples) and methodological concerns, such as reproducibility and robustness should be maximal; (c) check suitability of selected physiological biomarkers and their temporal integration for the targeted species and the processes studied; (d) select the most relevant biomarkers to avoid redundancy and ensure reasonable cost and time investments. This general approach is clearly transferable among species, locations and times.
For both management and conservation aspects, integrating the responses of multiple physiological biomarkers to different stressors can provide early warning signals of population responses to environmental changes and thereby serve as additional indicators of population state. Furthermore, the relationship between physiological biomarkers and fitness outcomes may help to refine long-term population projections through adjusted variability in growth, survival and connectivity parameters. By improving the accuracy of projections through physiological processes, we might be able to account for changes in life history traits and monitor the effects of environmental changes on exploited populations and thereby reduce the risks of collapse of these populations. To this end, collaboration between physiological ecologists, fisheries scientists, modellers and policy makers will be crucial to arm these biomarkers, pinpoint mechanisms underlying population declines, and set up appropriate management schemes facilitating the identification of future ecological problems much earlier than separate approaches of multiple disciplines alone. This will warrant meaningful and improved integration of both empirical and mechanistic understanding of physiology into existing and future models to address the management needs of many fish stocks.
References
Abdel-Tawwab M, Monier MN, Hoseinifar SH, Faggio C (2019) Fish response to hypoxia stress: growth, physiological, and immunological biomarkers. Fish Physiol Biochem 45:997–1013
Adams SM (1999) Ecological role of lipids in the health and success of fish populations. Lipids in freshwater ecosystems. Springer, New York, pp 132–160
Adams SM, Ham KD (2011) Application of biochemical and physiological indicators for assessing recovery of fish populations in a disturbed stream. Environ Manag 47:1047–1063. https://doi.org/10.1007/s00267-010-9599-7
Adams SM, Crumby WD, Greeley MS et al (1992) Relationships between physiological and fish population responses in a contaminated stream. Environ Toxicol Chem 11:1549–1557. https://doi.org/10.1002/etc.5620111105
Aerts J, Metz JR, Ampe B et al (2015) Scales tell a story on the stress history of fish. PLoS ONE. https://doi.org/10.1371/journal.pone.0123411
Ali M, Nicieza A, Wootton RJ (2003) Compensatory growth in fishes: a response to growth depression. Fish Fish 4:147–190. https://doi.org/10.1046/j.1467-2979.2003.00120.x
Baas J, Augustine S, Marques GM, Lou DJ (2018) Dynamic energy budget models in ecological risk assessment: from principles to applications. Sci Total Environ 628–629:249–260
Baker MR, Gobush KS, Vynne CH (2013) Review of factors influencing stress hormones in fish and wildlife. J Nat Conserv 21:309–318
Baskett ML (2012) Integrating mechanistic organism–environment interactions into the basic theory of community and evolutionary ecology. J Exp Biol 215:948–961. https://doi.org/10.1242/jeb.059022
Beauchard O, Veríssimo H, Queirós AM, Herman PMJ (2017) The use of multiple biological traits in marine community ecology and its potential in ecological indicator development. Ecol Indic 76:81–96
Beaulieu M, Costantini D (2014) Biomarkers of oxidative status: missing tools in conservation physiology. Conserv Physiol 2:1–16. https://doi.org/10.1093/conphys/cou014
Bergman JN, Bennett JR, Binley AD et al (2019) Scaling from individual physiological measures to population-level demographic change: case studies and future directions for conservation management. Biol Conserv. https://doi.org/10.1016/j.biocon.2019.108242
Bernos TA, Jeffries KM, Mandrak NE (2020) Linking genomics and fish conservation decision making: a review. Rev Fish Biol Fish 30:587–604
Birnie-Gauvin K, Costantini D, Cooke SJ, Willmore WG (2017) A comparative and evolutionary approach to oxidative stress in fish: a review. Fish Fish 18:928–942. https://doi.org/10.1111/faf.12215
Bonier F, Martin PR, Moore IT, Wingfield JC (2009) Do baseline glucocorticoids predict fitness? Trends Ecol Evol 24:634–642. https://doi.org/10.1016/j.tree.2009.04.013
Boshra H, Li J, Sunyer JO (2006) Recent advances on the complement system of teleost fish. Fish Shellfish Immunol 20:239–262. https://doi.org/10.1016/j.fsi.2005.04.004
Bozinovic F, Pörtner HO (2015) Physiological ecology meets climate change. Ecol Evol 5:1025–1030. https://doi.org/10.1002/ece3.1403
Breton AR, Hawkins JA, Winkelman DL (2013) Correcting length-frequency distributions for imperfect detection. N Am J Fish Manag 33:1156–1165. https://doi.org/10.1080/02755947.2013.829141
Brewster CL, Ortega J, Beaupre SJ (2020) Integrating bioenergetics and conservation biology: thermal sensitivity of digestive performance in Eastern Collared Lizards (Crotaphytus collaris) may affect population persistence. Conserv Physiol. https://doi.org/10.1093/conphys/coaa018
Brosset P, Smith AD, Plourde S et al (2020) A fine-scale multi-step approach to understand fish recruitment variability. Sci Rep. https://doi.org/10.1038/s41598-020-73025-z
Brown A, Thatje S (2014) Explaining bathymetric diversity patterns in marine benthic invertebrates and demersal fishes: physiological contributions to adaptation of life at depth. Biol Rev 89:406–426. https://doi.org/10.1111/brv.12061
Bueno-Pardo J, Petitgas P, Kay S, Huret M (2020) Integration of bioenergetics in an individual-based model to hindcast anchovy dynamics in the Bay of Biscay. ICES J Mar Sci 77:655–667. https://doi.org/10.1093/icesjms/fsz239
Burton T, Killen SS, Armstrong JD, Metcalfe NB (2011) What causes intraspecific variation in resting metabolic rate and what are its ecological consequences? Proc R Soc B Biol Sci 278:3465–3473
Cahu CL, Zambonino-Infante JL (1994) Early weaning of sea bass (Dicentrarchus labrax) larvae with a compound diet: effect on digestive enzymes. Comp Biochem Physiol Part A Physiol 109:213–222. https://doi.org/10.1016/0300-9629(94)90123-6
Catalán IA, Auch D, Kamermans P et al (2019) Critically examining the knowledge base required to mechanistically project climate impacts: a case study of Europe’s fish and shellfish. Fish Fish 20:501–517. https://doi.org/10.1111/faf.12359
Chabot D, McKenzie DJ, Craig JF (2016) Metabolic rate in fishes: definitions, methods and significance for conservation physiology. J Fish Biol 88:1–9
Cheng CHC, Detrich HW (2007) Molecular ecophysiology of Antarctic notothenioid fishes. Philos Trans R Soc B Biol Sci 362:2215–2232. https://doi.org/10.1098/rstb.2006.1946
Cheung WWL, Dunne J, Sarmiento JL, Pauly D (2011) Integrating ecophysiology and plankton dynamics into projected maximum fisheries catch potential under climate change in the Northeast Atlantic. ICES J Mar Sci 68:1008–1018. https://doi.org/10.1093/icesjms/fsr012
Clancey E, Byers JA (2014) The definition and measurement of individual condition in evolutionary studies. Ethology 120:845–854. https://doi.org/10.1111/eth.12272
Clark TD, Sandblom E, Jutfelt F (2013) Aerobic scope measurements of fishes in an era of climate change: respirometry, relevance and recommendations. J Exp Biol 216:2771–2782
Colin N, Porte C, Fernandes D et al (2016) Ecological relevance of biomarkers in monitoring studies of macro-invertebrates and fish in Mediterranean rivers. Sci Total Environ 540:307–323. https://doi.org/10.1016/j.scitotenv.2015.06.099
Connon RE, Jeffries KM, Komoroske LM et al (2018) The utility of transcriptomics in fish conservation. J Exp Biol. https://doi.org/10.1242/jeb.148833
Cooke SJ, O’Connor CM (2010) Making conservation physiology relevant to policy makers and conservation practitioners. Conserv Lett 3:159–166. https://doi.org/10.1111/j.1755-263X.2010.00109.x
Cooke SJ, Suski CD (2008) Ecological restoration and physiology: an overdue integration. Bioscience 58:957–968. https://doi.org/10.1641/B581009
Cooke SJ, Sack L, Franklin CE et al (2013) What is conservation physiology? Perspectives on an increasingly integrated and essential science. Conserv Physiol. https://doi.org/10.1093/conphys/cot001
Coristine LE, Robillard CM, Kerr JT et al (2014) A conceptual framework for the emerging discipline of conservation physiology. Conserv Physiol. https://doi.org/10.1093/conphys/cou033
Costantini D (2008) Oxidative stress in ecology and evolution: lessons from avian studies. Ecol Lett 11:1238–1251. https://doi.org/10.1111/j.1461-0248.2008.01246.x
Côté IM, Darling ES, Brown CJ (2016) Interactions among ecosystem stressors and their importance in conservation. Proc R Soc B Biol Sci 283:1–9
Cotter J, Mesnil B, Witthames P, Parker-Humphreys M (2009) Notes on nine biological indicators estimable from trawl surveys with an illustrative assessment for North Sea cod. Aquat Living Resour 22:135–153. https://doi.org/10.1051/alr/2009016
Couturier LIE, Michel LN, Amaro T et al (2020) State of art and best practices for fatty acid analysis in aquatic sciences. ICES J Mar Sci. https://doi.org/10.1093/icesjms/fsaa121
Cury PM, Christensen V (2005) Quantitative ecosystem indicators for fisheries management. ICES J Mar Sci 62:307–310. https://doi.org/10.1016/j.icesjms.2005.02.003
Denver RJ, Hopkins PM, McCormick SD et al (2009) Comparative endocrinology in the 21st century. Integr Comp Biol 49:339–348. https://doi.org/10.1093/icb/icp082
Depledge MH, Amaral-Mendes JJ, Daniel B et al (1993) The conceptual basis of the biomarker approach. In: Biomarkers, pp 15–29
Dhabhar FS (2002) Stress-induced augmentation of immune function—the role of stress hormones, leukocyte trafficking, and cytokines. Brain Behav Immun 16:785–798. https://doi.org/10.1016/S0889-1591(02)00036-3
Diaz Pauli B, Wiech M, Heino M, Utne-Palm AC (2015) Opposite selection on behavioural types by active and passive fishing gears in a simulated guppy Poecilia reticulata fishery. J Fish Biol 86:1030–1045. https://doi.org/10.1111/jfb.12620
Dupuy C, Galland C, Pichereau V et al (2015) Assessment of the European flounder responses to chemical stress in the English Channel, considering biomarkers and life history traits. Mar Pollut Bull 95:634–645. https://doi.org/10.1016/j.marpolbul.2014.11.033
Ernande B, Dieckmann U, Heino M (2004) Adaptive changes in harvested populations: plasticity and evolution of age and size at maturation. Proc Biol Sci 271:415–423. https://doi.org/10.1098/rspb.2003.2519
Fahd F, Yang M, Khan F, Veitch B (2021) A food chain-based ecological risk assessment model for oil spills in the Arctic environment. Mar Pollut Bull. https://doi.org/10.1016/j.marpolbul.2021.112164
Faroon O, Harris M, Llados F et al (2002) Toxicological profile for DDT, DDE, and DDD. In: ATSDR’s toxicological profiles, US Department of Health and Human Services, Atlanta, GA, USA
Farrell AP (1991) From Hagfish to Tuna: a perspective on cardiac function in fish. Physiol Zool 64:1137–1164. https://doi.org/10.1086/physzool.64.5.30156237
Farrell AP (2016) Pragmatic perspective on aerobic scope: peaking, plummeting, pejus and apportioning. J Fish Biol 88:322–343. https://doi.org/10.1111/jfb.12789
Farrell AP, Gallaugher PE, Routledge R (2001) Rapid recovery of exhausted adult coho salmon after commercial capture by troll fishing. Can J Fish Aquat Sci 58:2319–2324. https://doi.org/10.1139/f01-188
Faught E, Vijayan MM (2018) Maternal stress and fish reproduction: the role of cortisol revisited. Fish Fish 19:1016–1030. https://doi.org/10.1111/faf.12309
Gagliano M, McCormick MI (2009) Hormonally mediated maternal effects shape offspring survival potential in stressful environments. Oecologia 160:657–665. https://doi.org/10.1007/s00442-009-1335-8
Greenstreet SPR, Fraser HM, Rogers SI et al (2012) Redundancy in metrics describing the composition, structure, and functioning of the North Sea demersal fish community. ICES J Mar Sci 69:8–22. https://doi.org/10.1093/icesjms/fsr188
Gregory TR, Wood CM (1999) The effects of chronic plasma cortisol elevation on the feeding behaviour, growth, competitive ability, and swimming performance of juvenile rainbow trout. Physiol Biochem Zool 72:286–295. https://doi.org/10.1086/316673
Hamilton PB, Cowx IG, Oleksiak MF et al (2016) Population-level consequences for wild fish exposed to sublethal concentrations of chemicals—a critical review. Fish Fish 17:545–566. https://doi.org/10.1111/faf.12125
Hook SE, Gallagher EP, Batley GE (2014) The role of biomarkers in the assessment of aquatic ecosystem health. Integr Environ Assess Manag 10:327–341. https://doi.org/10.1002/ieam.1530
Horodysky AZ, Cooke SJ, Brill RW (2015) Physiology in the service of fisheries science: why thinking mechanistically matters. Rev Fish Biol Fish 25:425–447. https://doi.org/10.1007/s11160-015-9393-y
Huebert KB, Peck MA (2014) A day in the life of fish larvae: modeling foraging and growth using quirks. PLoS ONE. https://doi.org/10.1371/journal.pone.0098205
Hulbert AJ, Kelly MA, Abbott SK (2014) Polyunsaturated fats, membrane lipids and animal longevity. J Comp Physiol B Biochem Syst Environ Physiol 184:149–166. https://doi.org/10.1007/s00360-013-0786-8
Hunt KE, Innis C, Merigo C et al (2019) Ameliorating transport-related stress in endangered Kemp’s ridley sea turtles (Lepidochelys kempii) with a recovery period in saltwater pools. Conserv Physiol. https://doi.org/10.1093/conphys/coy065
Iwama GK, Thomas PT, Forsyth RB, Vijayan MM (1998) Heat shock protein expression in fish. Rev Fish Biol Fish 8:35–56. https://doi.org/10.1023/A:1008812500650
Jeffrey JD, Hasler CT, Chapman JM et al (2015) Linking landscape-scale disturbances to stress and condition of fish: implications for restoration and conservation. Integr Comp Biol 55:618–630. https://doi.org/10.1093/icb/icv022
Jeffries KM, Teffer A, Michaleski S et al (2021) The use of non-lethal sampling for transcriptomics to assess the physiological status of wild fishes. Comp Biochem Physiol Part B Biochem Mol Biol. https://doi.org/10.1016/j.cbpb.2021.110629
Jennings S (2005) Indicators to support an ecosystem approach to fisheries. Fish Fish 6:212–232. https://doi.org/10.1111/j.1467-2979.2005.00189.x
Jobling M, Johansen SJS (1999) The lipostat, hyperphagia and catch-up growth. Aquac Res 30:473–478. https://doi.org/10.1046/j.1365-2109.1999.00358.x
Johnstone CP, Lill A, Reina RD (2017) Use of erythrocyte indicators of health and condition in vertebrate ecophysiology: a review and appraisal. Biol Rev 92:150–168. https://doi.org/10.1111/brv.12219
Jørgensen C, Holt RE (2013) Natural mortality: Its ecology, how it shapes fish life histories, and why it may be increased by fishing. J Sea Res 75:8–18. https://doi.org/10.1016/j.seares.2012.04.003
Josrgensen EH, Johansen SJS, Jobling M (1997) Seasonal patterns of growth, lipid deposition and lipid depletion in anadromous Arctic charr. J Fish Biol 51:312–326. https://doi.org/10.1111/j.1095-8649.1997.tb01668.x
Katsanevakis S, Weber A, Pipitone C et al (2012) Monitoring marine populations and communities: methods dealing with imperfect detectability. Aquat Biol 16:31–52. https://doi.org/10.3354/ab00426
Killen SS (2014) Growth trajectory influences temperature preference in fish through an effect on metabolic rate. J Anim Ecol 83:1513–1522. https://doi.org/10.1111/1365-2656.12244
Killen SS, Marras S, Metcalfe NB et al (2013) Environmental stressors alter relationships between physiology and behaviour. Trends Ecol Evol 28:651–658. https://doi.org/10.1016/j.tree.2013.05.005
Kimball S, Gremer JR, Angert AL et al (2012) Fitness and physiology in a variable environment. Oecologia 169:319–329. https://doi.org/10.1007/s00442-011-2199-2
Kime DE, Nash JP, Scott AP (1999) Vitellogenesis as a biomarker of reproductive disruption by xenobiotics. Aquaculture 177:345–352. https://doi.org/10.1016/S0044-8486(99)00097-6
Kiron V, Fukuda H, Takeuchi T, Watanabe T (1995) Essential fatty acid nutrition and defence mechanisms in rainbow trout Oncorhynchus mykiss. Comp Biochem Physiol Part A Physiol 111:361–367. https://doi.org/10.1016/0300-9629(95)00042-6
Koenigstein S, Dahlke FT, Stiasny MH et al (2018) Forecasting future recruitment success for Atlantic cod in the warming and acidifying Barents Sea. Glob Change Biol 24:526–535. https://doi.org/10.1111/gcb.13848
Korte SM, Koolhaas JM, Wingfield JC, McEwen BS (2005) The Darwinian concept of stress: benefits of allostasis and costs of allostatic load and the trade-offs in health and disease. Neurosci Biobehav Rev 29:3–38. https://doi.org/10.1016/j.neubiorev.2004.08.009
Kotiaho JS (2002) Sexual selection and condition dependence of courtship display in three species of horned dung beetles. Behav Ecol 13:791–799. https://doi.org/10.1093/beheco/13.6.791
Laugen AT, Engelhard GH, Whitlock R et al (2014) Evolutionary impact assessment: accounting for evolutionary consequences of fishing in an ecosystem approach to fisheries management. Fish Fish 15:65–96. https://doi.org/10.1111/faf.12007
Lawrence MJ, Jain-Schlaepfer S, Zolderdo AJ et al (2018) Are 3 minutes good enough for obtaining baseline physiological samples from teleost fish? Can J Zool 96:774–786. https://doi.org/10.1139/cjz-2017-0093
Lawrence MJ, Raby GD, Teffer AK et al (2020) Best practices for non-lethal blood sampling of fish via the caudal vasculature. J Fish Biol 97:4–15. https://doi.org/10.1111/jfb.14339
Le Luyer J, Schull Q, Auffret P et al (2021) Dual RNAseq highlights the kinetics of skin microbiome and fish host responsiveness to bacterial infection. Anim Microbiome. https://doi.org/10.1186/s42523-021-00097-1
Lemieux H, Blier P, Dutil J-D (1999) Do digestive tract enzymes set a physiological limit on growth rate and food conversion efficiency in the Atlantic cod (Gadus morhua). Fish Physiol Biochem 20:293–303
Lennox RJ, Suski CD, Cooke SJ (2018) A macrophysiology approach to watershed science and management. Sci Total Environ 626:434–440
Lester RE, Close PG, Barton JL et al (2014) Predicting the likely response of data-poor ecosystems to climate change using space-for-time substitution across domains. Glob Change Biol 20:3471–3481. https://doi.org/10.1111/gcb.12634
Lie Ø, Evensen Ø, Sørensen A, Frøysadal E (1989) Study on lysozyme activity in some fish species. Dis Aquat Organ 6:1–5. https://doi.org/10.3354/dao006001
Link JS, Huse G, Gaichas S, Marshak AR (2020) Changing how we approach fisheries: a first attempt at an operational framework for ecosystem approaches to fisheries management. Fish Fish 21:393–434. https://doi.org/10.1111/faf.12438
Little AG, Loughland I, Seebacher F (2020) What do warming waters mean for fish physiology and fisheries? J Fish Biol 97:328–340. https://doi.org/10.1111/jfb.14402
Lloret J, Shulman G, Love RM (2014) Condition and health indicators of exploited marine fishes. Wiley, Oxford
Madliger CL, Love OP (2015) The power of physiology in changing landscapes: considerations for the continued integration of conservation and physiology. Integr Comp Biol 55:545–553. https://doi.org/10.1093/icb/icv001
Madliger CL, Cooke SJ, Crespi EJ et al (2016) Success stories and emerging themes in conservation physiology. Conserv Physiol 4:1–17. https://doi.org/10.1093/conphys/cov057
Madliger CL, Love OP, Hultine KR, Cooke SJ (2018) The conservation physiology toolbox: status and opportunities. Conserv Physiol 6:1–16. https://doi.org/10.1093/conphys/coy029
Mahoney JL, Klug PE, Reed WL (2018) An assessment of the US endangered species act recovery plans: using physiology to support conservation. Conserv Physiol. https://doi.org/10.1093/conphys/coy036
Mangel M, Levin PS (2005) Regime, phase and paradigm shifts: making community ecology the basic science for fisheries. Philos Trans R Soc B Biol Sci 360:95–105. https://doi.org/10.1098/rstb.2004.1571
Mangel M, Munch SB (2005) A life-history perspective on short- and long-term consequences of compensatory growth. Am Nat 166:155–176. https://doi.org/10.1086/444439
Martino EJ, Houde ED (2010) Recruitment of striped bass in Chesapeake Bay: spatial and temporal environmental variability and availability of zooplankton prey. Mar Ecol Prog Ser 409:213–228. https://doi.org/10.3354/meps08586
Maunder MN, Piner KR (2014) Contemporary fisheries stock assessment: many issues still remain. ICES J Mar Sci 72:7–18. https://doi.org/10.1093/icesjms/fsu015
McKenzie DJ, Axelsson M, Chabot D et al (2016) Conservation physiology of marine fishes: state of the art and prospects for policy. Conserv Physiol. https://doi.org/10.1093/conphys/cow046
Metcalfe NB, Monaghan P (2003) Growth versus lifespan: perspectives from evolutionary ecology. Exp Gerontol 38:935–940. https://doi.org/10.1016/S0531-5565(03)00159-1
Metcalfe JD, le Quesne WJF, Cheung WWL, Righton DA (2012) Conservation physiology for applied management of marine fish: an overview with perspectives on the role and value of telemetry. Philos Trans R Soc B Biol Sci 367:1746–1756
Metcalfe WS, Tudorache C, Wilson RP (2016) Recent advances in telemetry for estimating the energy metabolism of wild fishes. J Fish Biol 88:284–297. https://doi.org/10.1111/jfb.12804
Mochnacz NJ, Kissinger BC, Deslauriers D et al (2017) Development and testing of a simple field-based intermittent-flow respirometry system for riverine fishes. Conserv Physiol. https://doi.org/10.1093/conphys/cox048
Monaco CJ, Helmuth B (2011) Tipping points, thresholds and the keystone role of physiology in marine climate change research. In: Advances in marine biology, pp 123–160
Montero D, Kalinowski T, Obach A et al (2003) Vegetable lipid sources for gilthead seabream (Sparus aurata): effects on fish health. Aquaculture 225:353–370. https://doi.org/10.1016/S0044-8486(03)00301-6
Moyano M, Illing B, Polte P et al (2020) Linking individual physiological indicators to the productivity of fish populations: a case study of Atlantic herring. Ecol Indic. https://doi.org/10.1016/j.ecolind.2020.106146
Nash KL, Alexander K, Melbourne-Thomas J et al (2021) Developing achievable alternate futures for key challenges during the UN Decade of Ocean Science for Sustainable Development. Rev Fish Biol Fish. https://doi.org/10.1007/s11160-020-09629-5
Newsholme EA, Start C (1973) Regulation in metabolism
Nusslé S, Bornand CN, Wedekind C (2009) Fishery-induced selection on an Alpine whitefish: quantifying genetic and environmental effects on individual growth rate. Evol Appl 2:200–208. https://doi.org/10.1111/j.1752-4571.2008.00054.x
Parrish CC (2009) Essential fatty acids in aquatic food webs. In: Lipids in aquatic ecosystems, pp 309–326
Patterson DA, Cooke SJ, Hinch SG, Robinson KA, Young N, Farrell AP, Miller KM (2016) A perspective on physiological studies supporting the provision of scientific advice for the management of Fraser River sockeye salmon (Oncorhynchus nerka). Conser Physiol 4(1):cow026. https://doi.org/10.1093/conphys/cow026
Peck MA, Reglero P, Takahashi M, Catalán IA (2013) Life cycle ecophysiology of small pelagic fish and climate-driven changes in populations. Prog Oceanogr 116:220–245. https://doi.org/10.1016/j.pocean.2013.05.012
Pérez-Ruzafa A, Pérez-Marcos M, Marcos C (2018) From fish physiology to ecosystems management: keys for moving through biological levels of organization in detecting environmental changes and anticipate their consequences. Ecol Indic 90:334–345. https://doi.org/10.1016/j.ecolind.2018.03.019
Pickett STA (1989) Space-for-time substitution as an alternative to long-term studies. Long-term studies in ecology. Springer, New York, pp 110–135
Raposo De Magalhães C, Schrama D, Farinha AP et al (2020) Protein changes as robust signatures of fish chronic stress: a proteomics approach to fish welfare research. BMC Genomics. https://doi.org/10.1186/s12864-020-6728-4
Rice JC, Rochet MJ (2005) A framework for selecting a suite of indicators for fisheries management. ICES J Mar Sci 62:516–527. https://doi.org/10.1016/j.icesjms.2005.01.003
Ricklefs R, Wikelski M (2002) The physiology/life-history nexus. Trends Ecol Evol 17:462–468
Roberts RJ, Agius C, Saliba C et al (2010) Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis 33:789–801
Rochet MJ, Trenkel V, Bellail R et al (2005) Combining indicator trends to assess ongoing changes in exploited fish communities: diagnostic of communities off the coasts of France. ICES J Mar Sci 62:1647–1664. https://doi.org/10.1016/j.icesjms.2005.06.009
Rowe L, Houle D (1996) The lek paradox and the capture of genetic variance by condition dependent traits. Proc R Soc Ser B 263:1415–1421
Sadoul B, Vijayan MM (2016) Stress and growth. In: Fish physiology, pp 167–205
Sadoul B, Geffroy B (2019) Measuring cortisol, the major stress hormone in fishes. J Fish Biol 94:540–555. https://doi.org/10.1111/jfb.13904
Sargent J, McEvoy L, Estevez A et al (1999) Lipid nutrition of marine fish during early development: current status and future directions. Aquaculture 179:217–229. https://doi.org/10.1016/S0044-8486(99)00191-X
Scott GR, Sloman KA (2004) The effects of environmental pollutants on complex fish behaviour: integrating behavioural and physiological indicators of toxicity. Aquat Toxicol 68:369–392. https://doi.org/10.1016/j.aquatox.2004.03.016
Sheldon BC, Verhulst S (1996) Ecological immunology—costly parasite defenses and trade-offs in evolutionary ecology. Trends Ecol Evol 11:317–321. https://doi.org/10.1016/0169-5347(96)10039-2
Sierra-Flores R, Atack T, Migaud H, Davie A (2015) Stress response to anthropogenic noise in Atlantic cod Gadus morhua L. Aquac Eng 67:67–76. https://doi.org/10.1016/j.aquaeng.2015.06.003
Sies H (1997) Oxidative stress: oxidants and antioxidants. Exp Physiol 82:291–295
Šimková A, Lafond T, Ondračková M et al (2008) Parasitism, life history traits and immune defence in cyprinid fish from Central Europe. BMC Evol Biol. https://doi.org/10.1186/1471-2148-8-29
Skomal GB (2007) Evaluating the physiological and physical consequences of capture on post-release survivorship in large pelagic fishes. Fish Manag Ecol 14:81–89. https://doi.org/10.1111/j.1365-2400.2007.00528.x
Sloman KA, McNeil PL (2012) Using physiology and behaviour to understand the responses of fish early life stages to toxicants. J Fish Biol 81:2175–2198. https://doi.org/10.1111/j.1095-8649.2012.03435.x
Sogard SM (1997) Size-selective mortality in the juvenile stage of teleost fishes: a review. Bull Mar Sci 60:1129–1157
Sopinka NM, Donaldson MR, O’Connor CM et al (2016) Stress indicators in fish. In: Fish physiology, pp 405–462
Sopinka NM, Capelle PM, Semeniuk CAD, Love OP (2017) Glucocorticoids in fish eggs: variation, interactions with the environment, and the potential to shape offspring fitness. Physiol Biochem Zool 90:15–33. https://doi.org/10.1086/689994
Teal LR, Marras S, Peck MA, Domenici P (2018) Physiology-based modelling approaches to characterize fish habitat suitability: their usefulness and limitations. Estuar Coast Shelf Sci 201:56–63. https://doi.org/10.1016/j.ecss.2015.11.014
Thomas CD, Bodsworth EJ, Wilson RJ et al (2001) Ecological and evolutionary processes at expanding range margins. Nature 411:577–581. https://doi.org/10.1038/35079066
Thompson MSA, Pontalier H, Spence MA et al (2020) A feeding guild indicator to assess environmental change impacts on marine ecosystem structure and functioning. J Appl Ecol 57:1769–1781. https://doi.org/10.1111/1365-2664.13662
Timi JT, Poulin R (2003) Parasite community structure within and across host populations of a marine pelagic fish: how repeatable is it? Int J Parasitol 33:1353–1362. https://doi.org/10.1016/S0020-7519(03)00203-0
Tocher DR (2003) Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fish Sci 11:107–184
Tomkins JL, Radwan J, Kotiaho JS, Tregenza T (2004) Genic capture and resolving the lek paradox. Trends Ecol Evol 19:323–328. https://doi.org/10.1016/j.tree.2004.03.029
Tort L, Sunyer JO, Gómez E, Molinero A (1996) Crowding stress induces changes in serum haemolytic and agglutinating activity in the gilthead sea bream Sparus aurata. Vet Immunol Immunopathol 51:179–188. https://doi.org/10.1016/0165-2427(95)05502-9
Trana MR, Roth JD, Tomy GT et al (2015) Influence of sample degradation and tissue depth on blubber cortisol in beluga whales. J Exp Mar Biol Ecol 462:8–13. https://doi.org/10.1016/j.jembe.2014.10.010
Trenkel VM, Rochet MJ, Mesnil B (2007) From model-based prescriptive advice to indicator-based interactive advice. ICES J Mar Sci 64:768–774. https://doi.org/10.1093/icesjms/fsm006
Trenkel VM, Vaz S, Albouy C et al (2019) We can reduce the impact of scientific trawling on marine ecosystems. Mar Ecol Prog Ser 609:277–282. https://doi.org/10.3354/meps12834
Tuomainen U, Candolin U (2011) Behavioural responses to human-induced environmental change. Biol Rev 86:640–657. https://doi.org/10.1111/j.1469-185X.2010.00164.x
Ueberschär B (1995) The use of tryptic enzyme activity measurement as a nutritional condition index: laboratory calibration data and field application. Ices Mar Sci Symp 201:119–129
Vanderplancke G, Claireaux G, Quazuguel P et al (2014) Exposure to chronic moderate hypoxia impacts physiological and developmental traits of European sea bass (Dicentrarchus labrax) larvae. Fish Physiol Biochem 41:233–242. https://doi.org/10.1007/s10695-014-0019-4
Vinagre C, Madeira D, Narciso L et al (2012) Effect of temperature on oxidative stress in fish: Lipid peroxidation and catalase activity in the muscle of juvenile seabass, Dicentrarchus labrax. Ecol Indic 23:274–279. https://doi.org/10.1016/j.ecolind.2012.04.009
Viney ME, Riley EM, Buchanan KL (2005) Optimal immune responses: immunocompetence revisited. Trends Ecol Evol 20:665–669. https://doi.org/10.1016/j.tree.2005.10.003
Von Bertalanffy L (1957) Quantitative laws in metabolism and growth. Q Rev Biol 32:217–231. https://doi.org/10.1086/401873
Watson JW, Hyder K, Boyd R et al (2020) Assessing the sublethal impacts of anthropogenic stressors on fish: an energy-budget approach. Fish Fish 21:1034–1045. https://doi.org/10.1111/faf.1248
Walker BG, Boersma PD, Wingfield JC (2005) Field endocrinology and conservation biology. Integr Comp Biol 45:12–18. https://doi.org/10.1093/icb/45.1.12
Ward TD, Algera DA, Gallagher AJ et al (2016) Understanding the individual to implement the ecosystem approach to fisheries management. Conserv Physiol. https://doi.org/10.1093/conphys/cow005.10.1093/conphys/cow005
Wernberg T, Smale DA, Thomsen MS (2012) A decade of climate change experiments on marine organisms: procedures, patterns and problems. Glob Change Biol 18:1491–1498. https://doi.org/10.1111/j.1365-2486.2012.02656.x
Wikelski M, Cooke SJ (2006) Conservation physiology. Trends Ecol Evol 21:38–46
Wilson KL, Honsey AE, Moe B, Venturelli P (2018) Growing the biphasic framework: techniques and recommendations for fitting emerging growth models. Methods Ecol Evol 9:822–833. https://doi.org/10.1111/2041-210X.12931
Young JL, Bornik ZB, Marcotte ML et al (2006) Integrating physiology and life history to improve fisheries management and conservation. Fish Fish 7:262–283. https://doi.org/10.1111/j.1467-2979.2006.00225.x
Yu BP (1994) Cellular defenses against damage from reactive oxygen species. Physiol Rev 74:139–162. https://doi.org/10.1152/physrev.1994.74.1.139
Zambonino-Infante JL, Cahu CL (2010) Effect of nutrition on marine fish development and quality. In: Recent advances in aquaculture research, pp 103–124
Zambonino-Infante J, Gisbert E, Sarasquete C et al (2008) Ontogeny and physiology of the digestive system of marine fish larvae. In: Feeding and digestive functions in fishes
Zuk M, Stoehr AM (2002) Immune defense and host life history. Am Nat 160:9–22. https://doi.org/10.1086/342131
Acknowledgements
The authors are grateful to J-L. Zambonino-Infante for his feedback on previous versions of this manuscript, to Sébastien Hervé for artwork. P.B. was supported by ISblue project, Interdisciplinary graduate school for the blue planet (ANR-17-EURE-0015) and co-funded by a grant from the French government under the program "Investissements d'Avenir", and by a grant from the Regional Council of Brittany (SAD programme). We are also grateful to several anonymous referees for providing thoughtful comments on the manuscript.
Funding
P.B. was supported by ISblue project, Interdisciplinary graduate school for the blue planet (ANR-17-EURE-0015) and co-funded by a grant from the French government under the program "Investissements d'Avenir", and by a grant from the Regional Council of Brittany (SAD programme).
Author information
Authors and Affiliations
Contributions
PB and CL conceived the study. PB performed most of the literature search to which all co-authors contributed. The first draft of the manuscript was written by PB and CL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brosset, P., Cooke, S.J., Schull, Q. et al. Physiological biomarkers and fisheries management. Rev Fish Biol Fisheries 31, 797–819 (2021). https://doi.org/10.1007/s11160-021-09677-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-021-09677-5