Abstract
Traditional “Green Revolution” cereal breeding strategies to improve yield are now reaching a plateau in our principal global food crop rice. Photosynthesis has now become a major target of international consortia to increase yield potential. Synthetic biology is being used across multiple large projects to improve photosynthetic efficiency. This review follows the genesis and progress of one of the first of these consortia projects, now in its 13th year; the Bill and Melinda Gates funded C4 Rice Project. This project seeks to install the biochemical and anatomical attributes necessary to support C4 photosynthesis in the C3 crop rice. Here we address the advances made thus far in installing the biochemical pathway and some of the key targets yet to be reached.
Similar content being viewed by others
Photosynthesis and food security
The global population has now passed 8 billion and is predicted to reach more than 9 billion by the year 2040. Much has been written about our lack of capacity to feed this burgeoning population, with declining arable land areas, climate change, extreme weather events and stagnating progress in yields of our major cereal crops (UNICEF 2022; Furbank et al. 2020). While statistics such as the need for a 70% increase in food production by 2050 are sobering, the focus on the future can result in the lack of political will to address the issue with immediacy (WorldBank 2022). It has been suggested that the global food crisis, unlike COVID 19, is a silent pandemic, not causing the level of global response required to resolve the problem. Recent events comprising extreme and cataclysmic weather, rapidly rising fuel costs, supply chain issues, war and socioeconomic unrest have reminded us that the equation where food demand outstrips supply is near at hand. Indeed, we now appear to be revisiting the food crisis of 2008 which saw rapidly escalating food prices following the exhaustion of dwindling global grain reserves, resulting in famine and social unrest (FAO 2009).
While the causes of declining global food security are manifest, it is now widely accepted that the strategies used to achieve the huge “Green Revolution” gains in cereal grain productivity and land use gains (40–60% in wheat; Vietmeyer 2011; Stevenson et al. 2013) have largely been exhausted. Yield gains from reducing investment of fixed carbon into unproductive biomass such as stems via introduction of dwarfing genes, breeding for grain number and harvest index (the proportion of crop biomass comprising harvestable grain) appear to have plateaued (Furbank et al. 2020). As grain yield is a product of this “harvest index” and final crop biomass, increasing biomass through increases in photosynthetic efficiency have been identified as the next critical breeding target. Rice breeding has led the way for the global push to improve crop photosynthesis by utilizing an engineering approach based on basic scientific research, physiological knowledge and modelling.
Rice is limited by photosynthate supply
As early as the 1990’s, while debate as to the relationship between photosynthetic performance and yield was ongoing in other crops, in rice it was becoming clear that rice yields were limited by the supply of photosynthate (Sheehy et al. 2001). In the decade leading up to this realization, intensive breeding for a “new plant type” had been ongoing at the International Rice Research Institute (IRRI) in the Philippines, in China and elsewhere with less than encouraging results in some cases. Rice bred for high spikelet number actually produced less than 50% of the predicted yield, with less than half the juvenile spikelet’s reaching maturity as filled grain (Fig. 1A and Sheehy et al. 2001). Highly successful breeding for improvements in panicle spikelet number could in some cases even reduced the number of final filled grain, presumably by diluting the available photosynthate to the point that fertility was reduced and abortion increased. This was taken as clear evidence that high yield potential rice is “source limited” i.e. limited by the provision of photoassimilates to support grain filling. Similar source limitation is not seen in other crops such as wheat, where there is significant genetic variation in photosynthetic capacity across historic collections and elite material (Silva-Pérez et al. 2019), and photosynthetic supply has increased in concert with more gradual gains in sink demand (Fischer et al. 1998). We now know that in a number of rice breeding programs, that the step change in spikelet number was achieved due to a mutation of an enzyme of the cytokinin catabolism, cytokinin oxidase (Ashikari et al. 2005; Fig. 1B), resulting in sink demand outstripping carbon supply from leaves. Thus, in rice the demand and potential for large yield gains is present, and efforts to fill these additional spikelets has become a priority activity in rice breeding programs around the world.
Spikelet numbers for indica rice variety IR72 averaged across 1997 and 1999 growing season data at the International Rice Research Station Los Banos Philippines (IRRI). From Sheehy et al (2001). 1000 grain weight in IR72 was 24 g in both seasons. Figure 1B shows typical panicles from Koshikari and Habataki; parents used to generate a QTL mapping population for grain number by Ashikari et al (2005) wherein a polymorphism in the cytokinin oxidase gene was identified as a major causative QTL for grain number. Line 5150 is a high yielding variety from China shown to have a deletion at the cytokinin oxidase locus resulting in a null mutation. These authors also phenocopied this large panicle architecture by producing a transgenic rice where cytokinin oxidase expression was suppressed
Redesigning rice photosynthesis to increase yield: the genesis of the C4 rice program
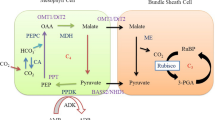
John Sheehy, a crop modeler working with physiological breeders at IRRI, recognized that photosynthetic improvement was key to boosting rice yields and brought together an international group of plant biologists to IRRI at the end of 1999 to brainstorm the problem. While a multitude of ideas were discussed (Sheehy et al. 2000), Sheehy’s favourite option was a bold and ambitious plan to use genetic technologies to introduce into rice the entire C4 photosynthetic pathway, including anatomical specialization. He had calculated from modelling that this alone, among the options discussed, could improve radiation use efficiency to the level required to produce a 50% yield boost; the improvement required to meet projected demand in 2050 (Sheehy et al. 2000). In most species the C4 pathway is a complex combination of both biochemical and morphological specialisation, which provides an elevation of the CO2 concentration at the site of Rubisco in the bundle sheath. The C4 cycle, often called a biochemical CO2 pump, fixes CO2 into C4 acids in the mesophyll via phosphoenolpyruvate carboxylase (PEPC) which diffuse to and are decarboxylated in the bundle sheaths allowing CO2 to be concentrated there (Fig. 2; von Caemmerer and Furbank 2003).
Schematic of enzymes and transporters included in the construction of C4 rice (in blue). These enzymes included carbonic anhydrase (CA), PEP carboxylase (PEPC), malate dehydrogenase (MDH), NADP-malic enzyme (NADP-ME), and pyruvate phosphate dikinase (PPDK). The transporters included are oxaloacetate/malate translocator (OMT1), pyruvate/sodium symporter (BASS2), Sodium/proton antiporter (NDH1), and PEP/phosphate translocator (PTT1). In bundle sheath chloroplasts it is unknown what is counter exchanged for malate or how pyruvate is exported. Also shown is the possible triose phosphate shuttle of the C3 cycle between bundle sheath and mesophyll cells. PYR pyruvate, PEP phosphoenolpyruvate, OAA oxaloacetate, MAL malate, 3PGA 3-phosphoglycerate, 1,3BPG 1,3-bisphosphoglycerate, G3P glycerate-3-phosphate, DHAP dihydroxyacetone phosphate. TPT triose-phosphate/phosphate translocator, PGK phosphoglycerate kinase, GAPDH glyceraldehyde-3-phosphate dehydrogenase, TPI triosephosphate isomerase
At the time of this meeting, some progress had already been made to express several maize photosynthetic genes in rice leaves with the goal to produce C4 photosynthesis in a single cell, where carboxylation by PEPC and decarboxylation by NADP-malic enzyme (NADP-ME) both occurred in mesophyll cells but partitioned between the cytosol and chloroplast (Miyao et al. 2011; Miyao 2003). While it was unknown whether such a small spatial separation of the “CO2 pump” from Rubisco would be sufficient for the mechanism to function efficiently, this approach did not require the challenges of anatomical specialization (Miyao et al. 2011). Initial promising reports pointed toward yield and growth increases with a partial pathway installed (Ku et al. 2000) later proving difficult to replicate and no functional C4 photosynthetic flux was ever demonstrated (Fukayama et al. 2003).
In 2006, at a follow up meeting at IRRI, the C4 Rice Consortium was born with a group of 16 laboratories in 11 countries resolving to take on the challenge of engineering C4 rice. The proceedings of this meeting (Sheehy et al. 2007) laid out the ground work for what was to become the most challenging crop engineering project ever attempted; what we would now call a major challenge in synthetic biology. Over the following 2 years, project plans were painstakingly prepared, timelines of ≥ 15 years for a prototype proposed and in 2008 a group of scientists representing the consortium presented the project to the Bill and Melinda Gates Foundation in Seattle. The backdrop to this presentation was what is now known as the global food crisis of 2008; a major deficit in global food supplies, the doubling of rice grain prices in less than a year, and severe shortages of wheat grain resulting in starvation and food riots in Cairo (FAO 2009). A perfect storm to support the funding of what many researchers called an “Apollo project”; as difficult as putting a man on the moon.
Building the tools to construct the prototype
C4 photosynthesis has evolved independently more than 65 times in nature (Sage et al. 2012). This has been achieved using three major biochemical variants of the pathway; NADP-malic enzyme (NADP-ME), NAD-ME malic (NAD-ME) and the Pepcarboxykinase (PCK) type (Furbank 2011), so named for the bundle sheath enzyme used to decarboxylate the C4 acid produced in the mesophyll cells to release CO2. Most cultivated C4 crops (such as maize, sorghum and sugarcane) utilize NADP-ME as their primary decarboxylase, leading the consortium to choose maize as the model for the C4 Rice prototype. Figure 2 shows the basic requirements of a C4 mechanism in rice including the key enzymes and chloroplast transport steps. Previous reviews have dealt in depth with the biochemical and anatomical specialization in C4 plants believed to have evolved to support the photosynthetic mechanism (von Caemmerer et al. 2012; Langdale 2011; Furbank 2011; Hibberd et al. 2008; Wang et al. 2017). These include reduced vein spacing with Kranz anatomy/photosynthetic functionalization of the bundle sheath (Wang et al. 2017; Langdale 2011), more than 100-fold increases in expression of key photosynthetic enzymes, and appropriate cell preferential expression of proteins in the bundle sheath or mesophyll (Ermakova et al. 2020). Here we will address progress and challenges in building tools to address two key aspects of building the C4 rice prototype; high level expression of pathway enzymes and appropriate cell preferential expression in mesophyll or bundle sheath compartments of rice (Ermakova et al. 2020).
Boosting C4 enzyme levels in rice; how much is enough?
An aspirational goal in C4 rice engineering is to reach maize levels of expression and/or activity of C4 enzymes preferentially expressed in the correct cell types, in a rice genotype with maize leaf anatomy. Although both maize and sorghum bicolor have exceptional high photosynthetic rates and enzyme activities, other C4 NADP-ME species such as Setaria viridis demonstrate that functional NADP-ME C4 photosynthesis can be achieved with a diverse range of enzyme activities (Sonawane et al. 2017; Osborn et al. 2017), mirroring the phylogenetic diversity of biochemical solutions observed across C4 species.
In addition to being guided by natural diversity in C4 pathways, we can also learn from previous work in transgenic C4 plants where levels of key enzymes were “titrated out” using gene suppression, as to the need to reach this aspirational goal in order to support adequate C4 photosynthetic fluxes in rice (Furbank et al. 1997). It can be seen in Fig. 2 that five C4 cycle enzymes support a minimal pathway (carbonic anhydrase (CA), PEP carboxylase (PEPC), malate dehydrogenase (MDH), NADP-malic enzyme (NADP-ME), and pyruvate phosphate dikinase (PPDK)). These enzymes have all been expressed in rice using a single construct but the amount of protein and enzyme activities need to be augmented (Ermakova et al. 2021). In Table 1 we have collated results from previous transgenic studies that have used antisense or RNAi technology to reduce the protein content of the C4 enzymes. It summarizes the affect a 50% reduction in enzyme content would have on photosynthetic rate. Three studies have confirmed that there is ample carbonic anhydrase activity such that a 50% reduction results in a very small decrease in photosynthetic rate. Studies in Amaranthus edulis and Setaria viridis show that 45% PEPC activity leads to 21% reduction in rate highlighting the importance of this enzyme. Reduction in, (PPDK) by 45% also leads to a 17% reduction. Reduction in MDH activity by half does not reduce photosynthetic rate and similarly, reduction of malic enzyme activity by 50% results in only a 6% reduction in photosynthetic rate. This is encouraging and suggests that less than full maize enzyme activity levels may be able to produce a functional C4 rice. Rubisco located in the bundle sheath can exert the strongest control although it is often co-limited by electron capacity (Furbank et al. 1996; Siebke et al. 1997). Balancing the amount of Rubisco with the C4 enzyme actives will be an essential fine tuning to ensure the efficiency of the pathway (von Caemmerer et al. 1997a, b).
The concentration of CO2 around Rubisco in the bundle sheath cells of C4 plants is dependent on unidirectional flow of metabolites through the C4 pathway (Fig. 2). Achieving this unidirectional flow is critically dependent on tight cell type specific regulation of enzyme and transporter activity to limit futile cycling of C4 cycle metabolites. Although there are multiple ways in which cell type specific activity could be achieved (e.g. through a combination of transcriptional and/or post-translational processes), analysis of cell type specific transcriptomes revealed that regulation at the level of gene transcription has been the primary way in which cell type specificity has been achieved in different C4 species (John et al. 2014; Emms et al. 2016; Chang et al. 2012). The genes encoding the enzymes and transporters of the C4 cycle show extreme differences in transcript abundance between bundle sheath and mesophyll cells (Fig. 3), while the orthologous genes in rice show little (Fig. 3; Hua et al. 2021). Thus, the aim of the C4 rice project has been to achieve high levels of cell-type specific enzyme activity through the use of cell type specific promoters to drive expression of exogenous transgenes.
Transcript abundance of genes encoding enzymes and transporters of the C4 cycle. Bundle sheath and mesophyll cell transcript abundance for Oryza sativa, Setaria viridis, Zea mays and Sorghum bicolor. All abundance estimates are provided as transcripts per million (TPM). For abbreviations of gene names see Fig. 2
Synthetic biology accelerates progress
A research project which has been working toward a single goal for more than a decade is a rarity in plant science. In the course of 13 years of work, advances in synthetic biology have revolutionized pathway engineering in plants. Hierarchical Golden Gate/MoClo or Golden Braid cloning (Engler et al. 2014; Andreou and Nakayama 2018), coupled with affordable gene synthesis means that rather than creating single gene transgenic plants and crossing, the tools are now available to build gene constructs containing many genes of interest and install them in a single step (see Fig. 4 and Ermakova et al. 2020). This means that genes of interest are inserted at a single genetic locus, can easily be tracked in subsequent crossing, and homozygous individuals can be generated without many years of successive crossing and screening for plants homozygous in all transgenic insertions (Lin et al. 2020; Fig. 4). Furthermore, the parts required for gene construction can be assembled in a “toolbox” for later use with other genes of interest or refining of the approach swapping promoters, introns, untranslated and coding regions. The transgene expression challenges outlined above concerning high level and cell preferential expression are also addressed with new synthetic biology tools.
Schematic of a traditional crossing strategy to stack 5 C4 pathway related genes in rice similar to that used in Lin et al (2020) compared to the Golden Gate assembly method used in Ermakova et al (2020) for 5 genes using a single gene construct. The former process requires approximately 6 years of crossing and large scale genotyping due to the necessity to obtain individual lines for crossing expressing each enzymes at desired levels and homozygosity for all transgenes which will have inserted at different loci. In contrast single gene insertion means that once lines expressing all gens on the t-DNA at high levels have been obtained, they can be selfed to produce homozygous progeny in less than 1 year. Abbreviations as for Fig. 2. AGDC signifies transgenic lines where glycine decarboxylase expression has been suppressed in the mesophyll compartment
Cell specific expression of C4 proteins in rice has been a challenge due to the paucity of promoters known to express in the bundle sheath compartment (Ermakova et al. 2020). Until recently, a single plant promoter was available which showed bundle sheath cell preferential expression in rice; the Zoysia japonica PCK promoter (Nomura et al. 2005). It can be seen from Fig. 4 that several genes are required to be expressed in the bundle sheath compartment, requiring this single promoter to be re-used. There is considerable evidence in this project and elsewhere that repeating a promoter sequence in transgenes commonly results in problems with cloning due to recombination deletion or with in planta methylation and inactivation (Wassenegger 2002).
We recently demonstrated that the synthetic transcription activator-like effector (dTALE)/synthetic TALE-activated promoter (STAP) system (Brückner et al. 2015; Danila et al. 2022) provides a potential solution to the problems outlined above (Fig. 5). This system, adapted from the plant immune response to bacterial infection (Bogdanove et al. 2010), allows multiplexing of several genes on a single construct where each gene of interest is driven by a different STAP, but all STAPs are activated by a single trans activating factor or dTALE (Schreiber and Tissier 2017). In the case of C4 engineering, this means that a single cell preferential plant promoter can be used to drive a dTALE which activates a suite of STAPs, each driving a gene of interest in the cellular compartment where the dTALE is expressed (Fig. 5). An added advantage of this system is that it has been observed that the dTALE-STAP system can substantially amplify the strength of a weak promoter (Danila et al. 2022), potentially also addressing the problem of sufficient high-level expression in the correct cell type in the C4 rice prototype.
Cell specific multiplexing strategy using synthetic promoters and transcriptional activators (Danila et al 2022). Two separate dTALE transitional activators are used, each capable of activating as set of STAP synthetic promoters. In one case (dTALE1), a mesophyll promoter is used to express the transitional activator only in this compartment hence activating expression of the multiplexed transgene set in this cell type only. In the other case (dTALE2), a bundle sheath specific promoter is used to provide activation of expression in a similar way. This approach avoids the re-use of promoter sequences and the necessity to have several plant promoters specific to the target cell type
Future challenges
The synthetic promoter system described above can potentially address the high-level expression in the correct compartment of the C4 enzymes in the basic pathway. “Maize levels” of every enzyme and transporter may be unnecessary to achieve appropriate flux as discussed above (Table 1). However, estimating flux through individual steps of the pathway is challenging. While extractable activity may be high, in vivo activity could be limited by inappropriate regulation or in fact substrate supply (Fukayama et al. 2003). The success of engineering a complete pathway can only really be judged by measurements of flux, such as via labelling into C4 acids and subsequent release and refixation of this carbon into the C3 cycle by Rubisco (Ermakova et al. 2021). If flux is low, trouble shooting of components in a complex prototype which includes multiple enzymes and transporters is challenging since localization of labelled metabolites in particular cell types or subcellular compartments is extremely difficult. Even if moderate fluxes are achieved, in the absence of the appropriate Kranz anatomy, the C4 pathway can likely only operate in cells adjacent to the bundle sheath cells (Ermakova et al. 2020). Vascular bundles in rice are separated by 6–9 mesophyll cells, all carrying out C3 photosynthesis, while only 2 mesophyll cells, devoid of Rubisco, are present between bundle sheath cells in the majority of C4 leaves (Ermakova et al. 2020; Sedelnikova et al. 2018). While modelling predicts measurable physiological impacts on gas exchange if maize levels of flux were achieved around the veins in rice (Ermakova et al. 2020), measuring the passage of label through C4 acids and then into 3-phosphoglycerate will be challenging to detect against the large background of labelled carbon appearing in 3-C compounds by direct fixation via Rubisco in mesophyll cells. For these technical reasons alone, it is desirable to combine close vein spacing and/or low Rubisco in the mesophyll tissues in subsequent prototypes. While some progress has been made on photosynthetic functionalization of the bundle sheath cells in rice (Wang et al. 2017; Ermakova et al. 2020), a major challenge remains; the discovery of a complete set of genetic switches required for the transition from C3 to C4 leaf vein spacing which remain elusive (Sedelnikova et al. 2018).
Concluding comments
The C4 Rice project is progressing on several fronts. While there remain a number of unanswered questions around transport of metabolites and genes controlling vein spacing and anatomy (Wang et al. 2016; Ermakova et al. 2020), modelling suggests that even without a full complement of anatomical specialisation, we can achieve a boost in photosynthesis and yield (Ermakova et al. 2020). The rate of technological progress in plant pathway engineering and synthetic biology since the genesis of the project provides hope that we can quickly deploy scientific discoveries into our prototype, even fine tuning with rapid advances in crop gene editing, to assure success.
References
Andreou AI, Nakayama N (2018) Mobius Assembly: A versatile Golden-Gate framework towards universal DNA assembly. Plos One 13(1):e0189892. https://doi.org/10.1371/journal.pone.0189892
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309(5735):741–745. https://doi.org/10.1126/science.1113373
Bogdanove AJ, Schornack S, Lahaye T (2010) TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol 13(4):394–401. https://doi.org/10.1016/j.pbi.2010.04.010
Brückner K, Schäfer P, Weber E, Grützner R, Marillonnet S, Tissier A (2015) A library of synthetic transcription activator-like effector-activated promoters for coordinated orthogonal gene expression in plants. Plant J 82(4):707–716. https://doi.org/10.1111/tpj.12843
Chang Y-M, Liu W-Y, Shih AC-C, Shen M-N, Lu C-H, Lu M-YJ, Yang H-W, Wang T-Y, Chen SC-C, Chen SM, Li W-H, Ku MSB (2012) Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160(1):165–177. https://doi.org/10.1104/pp.112.203810
Cousins AB, Baroli I, Badger MR, Ivakov A, Lea PJ, Leegood RC, von Caemmerer S (2007) The role of phosphoenolpyruvate carboxylase during C4 photosynthetic isotope exchange and stomatal conductance. Plant Physiol 145(3):1006–1017
Danila F, Schreiber T, Ermakova M, Hua L, Vlad D, Lo S-F, Chen Y-S, Lambret-Frotte J, Hermanns AS, Athmer B, von Caemmerer S, Yu S-M, Hibberd JM, Tissier A, Furbank RT, Kelly S, Langdale JA (2022) A single promoter-TALE system for tissue-specific and tuneable expression of multiple genes in rice. Plant Biotechnol J 20(9):1786–1806. https://doi.org/10.1111/pbi.13864
Emms DM, Covshoff S, Hibberd JM, Kelly S (2016) Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol Biol Evol 33(7):1796–1806. https://doi.org/10.1093/molbev/msw057
Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3(11):839–843. https://doi.org/10.1021/sb4001504
Ermakova M, Danila FR, Furbank RT, von Caemmerer S (2020) On the road to C4 rice: advances and perspectives. Plant J 101(4):940–950. https://doi.org/10.1111/tpj.14562
Ermakova M, Arrivault S, Giuliani R, Danila F, Alonso-Cantabrana H, Vlad D, Ishihara H, Feil R, Guenther M, Borghi GL, Covshoff S, Ludwig M, Cousins AB, Langdale JA, Kelly S, Lunn JE, Stitt M, von Caemmerer S, Furbank RT (2021) Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnol J 19(3):575–588. https://doi.org/10.1111/pbi.13487
FAO (2009) How to feed the world: global agriculture towards 2050. www.faoorg/fileadmin/templates/wsfs/docs/Issues_papers/HLEF2050_Global_Agriculturepdf
Fischer RA, Rees D, Sayre KD, Lu Z-M, Condon AG, Saavedra AL (1998) Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Sci. https://doi.org/10.2135/cropsci1998.0011183X003800060011x
Fukayama H, Hatch MD, Tamai T, Tsuchida H, Sudoh S, Furbank RT, Miyao M (2003) Activity regulation and physiological impacts of maize C-4-specific phosphoenolpyruvate carboxylase overproduced in transgenic rice plants. Photosynth Res 77(2–3):227–239
Furbank RT (2011) Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J Exp Bot 62(9):3103–3108
Furbank RT, Chitty JA, von Caemmerer S, Jenkins CLD (1996) Antisense RNA inhibition of RbcS gene expression reduces rubisco level and photosynthesis in the C4 plant Flaveria bidentis. Plant Physiol 111(3):725–734
Furbank RT, Chitty JA, Jenkins CLD, Taylor WC, Trevanion SJ, von Caemmerer S, Ashton AR (1997) Genetic manipulation of key photosynthetic enzymes in the C4 plant Flaveria bidentis. Aust J Plant Physiol 24(4):477–485
Furbank RT, Sharwood R, Estavillo GM, Silva-Perez V, Condon AG (2020) Photons to food: genetic improvement of cereal crop photosynthesis. J Exp Bot. https://doi.org/10.1093/jxb/eraa077
Hibberd JM, Sheehy JE, Langdale JA (2008) Using C4 photosynthesis to increase the yield of rice - rationale and feasibility. Curr Opin Plant Biol 11(2):228–231
Hua L, Stevenson SR, Reyna-Llorens I, Xiong H, Kopriva S, Hibberd JM (2021) The bundle sheath of rice is conditioned to play an active role in water transport as well as sulfur assimilation and jasmonic acid synthesis. Plant J 107(1):268–286. https://doi.org/10.1111/tpj.15292
John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM (2014) Evolutionary convergence of cell-specific gene expression in independent lineages of C-4 grasses. Plant Physiol 165(1):62–75
Ku MSB, Ranade U, Hsu TP, Cho D, Li X, Jiao DM, Ehleringer J, Miyao M, Matsuoka M (2000) Photosynthetic performance of transgenic rice plants overexpressing maize C4 photosynthesis enzymes**Citation: Sheehy JE, Mitchell PL, Hardy B, editors. 2000. Redesigning rice photosynthesis to increase yield. Proceedings of the Workshop on The Quest to Reduce Hunger: Redesigning Rice Photosynthesis, 30 Nov.-3 Dec. 1999, Los Bafios, Philippines. Makati City (Philippines): International Rice Research Institute and Amsterdam (The Netherlands): Elsevier Science B.V. 293 vol 7. Elsevier, pp. 193–204. https://doi.org/10.1016/S0928-3420(00)80015-4
Langdale JA (2011) C4 cycles: past, present, and future research on C4 photosynthesis [Review]. Plant Cell 23(11):3879–3892
Lin H, Arrivault S, Coe RA, Karki S, Covshoff S, Bagunu E, Lunn JE, Stitt M, Furbank RT, Hibberd JM, Quick WP (2020) A partial C4 photosynthetic biochemical pathway in rice. Front Plant Sci. https://doi.org/10.3389/fpls.2020.564463
Miyao M (2003) Molecular evolution and genetic engineering of C-4 photosynthetic enzymes [Review]. J Exp Bot 54(381):179–189
Miyao M, Masumoto C, Miyazawa S-I, Fukayama H (2011) Lessons from engineering a single-cell C4 photosynthetic pathway into rice. J Exp Bot 62(9):3021–3029. https://doi.org/10.1093/jxb/err023
Nomura M, Higuchi T, Katayama K, Taniguchi M, Miyao-Tokutomi M, Matsuoka M, Tajima S (2005) The promoter for C4-type mitochondrial aspartate aminotransferase does not direct Bundle sheath-specific expression in transgenic rice plants. Plant Cell Physiol 46(5):743–753. https://doi.org/10.1093/pcp/pci077
Osborn HL, Alonso-Cantabrana H, Sharwood RE, Covshoff S, Evans JR, Furbank RT, von Caemmerer S (2017) Effects of reduced carbonic anhydrase activity on CO2 assimilation rates in Setaria viridis: a transgenic analysis. J Exp Bot 68(2):299–310. https://doi.org/10.1093/jxb/erw357
Pengelly JJL, Tan J, Furbank RT, von Caemmerer S (2012) Antisense reduction of NADP-malic enzyme in Flaveria bidentis reduces flow of CO2 through the C4 Cycle. Plant Physiol 160(2):1070–1080
Sage RF, Sage TL, Kocacinar F (2012) Photorespiration and the evolution of C4 Photosynthesis. Annu Rev Plant Biol 63(1):19–47. https://doi.org/10.1146/annurev-arplant-042811-105511
Schreiber T, Tissier A (2017) Generation of dTALEs and Libraries of Synthetic TALE-activated promoters for engineering of gene regulatory networks in plants. In: Kaufmann K, Mueller-Roeber B (eds) Plant Gene regulatory networks: Methods and protocols. Springer New York, New York, pp 185–204
Sedelnikova OV, Hughes TE, Langdale JA (2018) Understanding the genetic basis of C4 Kranz anatomy with a view to engineering C3 Crops. Annu Rev Genet 52(1):249–270. https://doi.org/10.1146/annurev-genet-120417-031217
Sheehy JE, Mitchell PL, Hardy B (2000) Redesigning rice photosynthesis to increase yield. Proc of the Workshop on The Quest to Reduce Hunger: Redesigning Rice Photosynthesis. 30 Nov-3Dec 1999 Elsevier Science, Los Banos Philippines, Makati City , Netherlands
Sheehy JE, Dionora MJA, Mitchell PL (2001) Spikelet numbers, sink size and potential yield in rice. Field Crop Res 71(2):77–85
Sheehy J, Mitchell AK, Hardy B (2007) Reconfiguring the Rice Plant’s Photosynthetic Pathway. IRRI, Philippines
Siebke K, von Caemmerer S, Badger M, Furbank RT (1997) Expressing an rbcs antisense gene in transgenic Flaveria bidentis leads to an increased quantum requirement for CO2 fixed in photosystems I and II. Plant Physiol 115(3):1163–1174
Silva-Pérez V, De Faveri J, Molero G, Deery DM, Condon AG, Reynolds MP, Evans JR, Furbank RT (2019) Genetic variation for photosynthetic capacity and efficiency in spring wheat. J Exp Bot 71(7):2299–2311. https://doi.org/10.1093/jxb/erz439
Sonawane BV, Sharwood RE, von Caemmerer S, Whitney SM, Ghannoum O, Raines C (2017) Short-term thermal photosynthetic responses of C4 grasses are independent of the biochemical subtype. J Exp Bot. https://doi.org/10.1093/jxb/erx350
Stevenson JR, Villoria N, Byerlee D, Kelley T, Maredia M (2013) Green Revolution research saved an estimated 18 to 27 million hectares from being brought into agricultural production. Proc Natl Acad Sci 110(21):8363–8368. https://doi.org/10.1073/pnas.1208065110
Studer AJ, Gandin A, Kolbe AR, Wang L, Cousins AB, Brutnell TP (2014) A limited role for carbonic anhydrase in C4 photosynthesis as revealed by a ca1ca2 double mutant in maize. Plant Physiol 165(2):608–617
Trevanion SJ, Furbank RT, Ashton AR (1997) NADP-malate dehydrogenase in the C4 plant Flaveria bidentis - cosense suppression of activity in mesophyll and bundle-sheath cells and consequences for photosynthesis. Plant Physiol 113(4):1153–1165
UNICEF (2022) The State of Food Security and Nutrition in the World 2022. Repurposing food and agricultural policies to make healthy diets more affordable
Vietmeyer N (2011) Our daily bread: the essential norman borlaug. Bracing Books, Lorton
von Caemmerer S, Furbank RT (2003) The C4 pathway: an efficient CO2 pump. Photosynth Res 77(2–3):191–207
von Caemmerer S, Ludwig M, Millgate A, Farquhar GD, Price D, Badger M, Furbank RT (1997a) Carbon isotope discrimination during C4 photosynthesis: Insights from transgenic plants. Aust J Plant Physiol 24(4):487–494
von Caemmerer S, Millgate A, Farquhar GD, Furbank RT (1997b) Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase by antisense RNA in the C4 plant Flaveria bidentis leads to reduced assimilation rates and increased carbon isotope discrimination. Plant Physiol 113(2):469–477
von Caemmerer S, Quinn V, Hancock NC, Price GD, Furbank RT, Ludwig M (2004) Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant Cell Environ 27(6):697–703
von Caemmerer S, Quick WP, Furbank RT (2012) The development of C4 rice: current progress and future challenges. Science 336(6089):1671–1672
Wang P, Vlad D, Langdale JA (2016) Finding the genes to build C4 rice. Curr Opin Plant Biol 31:44–50. https://doi.org/10.1016/j.pbi.2016.03.012
Wang P, Khoshravesh R, Karki S, Tapia R, Balahadia CP, Bandyopadhyay A, Quick WP, Furbank R, Sage TL, Langdale JA (2017) Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr Biol 27(21):3278-3287.e3276. https://doi.org/10.1016/j.cub.2017.09.040
Wassenegger M (2002) Gene silencing. In: Jeon KW (ed) International Review of Cytology, vol 219. Academic Press, Cambridge, pp 61–113
WorldBank (2022) World Bank Food security update October 13 2022.https://www.worldbank.org/en/topic/agriculture/brief/food-security-update
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing the manuscript text, consolidating published data and constructing all the figures.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Furbank, R., Kelly, S. & von Caemmerer, S. Photosynthesis and food security: the evolving story of C4 rice. Photosynth Res 158, 121–130 (2023). https://doi.org/10.1007/s11120-023-01014-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1007/s11120-023-01014-0