Abstract
Site-specific estimation of lime requirement requires high-resolution maps of soil organic carbon (SOC), clay and pH. These maps can be generated with digital soil mapping models fitted on covariates observed by proximal soil sensors. However, the quality of the derived maps depends on the applied methodology. We assessed the effects of (i) training sample size (5–100); (ii) sampling design (simple random sampling (SRS), conditioned Latin hypercube sampling (cLHS) and k-means sampling (KM)); and (iii) prediction model (multiple linear regression (MLR) and random forest (RF)) on the prediction performance for the above mentioned three soil properties. The case study is based on conditional geostatistical simulations using 250 soil samples from a 51 ha field in Eastern Germany. Lin’s concordance correlation coefficient (CCC) and root-mean-square error (RMSE) were used to evaluate model performances. Results show that with increasing training sample sizes, relative improvements of RMSE and CCC decreased exponentially. We found the lowest median RMSE values with 100 training observations i.e., 1.73%, 0.21% and 0.3 for clay, SOC and pH, respectively. However, already with a sample size of 10, models of moderate quality (CCC > 0.65) were obtained for all three soil properties. cLHS and KM performed significantly better than SRS. MLR showed lower median RMSE values than RF for SOC and pH for smaller sample sizes, but RF outperformed MLR if at least 25–30 or 75–100 soil samples were used for SOC or pH, respectively. For clay, the median RMSE was lower with RF, regardless of sample size.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Precision agriculture (PA) strives to increase crop yields and economic productivity while reducing negative environmental impacts through spatially optimized field management (Gebbers & Adamchuk, 2010; Stoorvogel et al., 2015). A key focus in precision agriculture is the variable-rate application of lime (VRL) because soil fertility is largely controlled by its acidity (Holland et al., 2018). VRL is important as the optimal lime requirement can change every few meters due to small-scale soil variability within a field (Vogel et al., 2022). The lime requirement is ideally calculated from soil pH, clay- and soil organic carbon (SOC) content to take the soil buffer capacity into account (Rühlmann et al., 2021; Söderström et al., 2016). Hence, high-resolution maps of pH, clay and SOC content are required for effective implementation of VRL. Numerous studies have demonstrated how combinations of different on-the-go proximal soil sensors can be used to create such high-resolution soil maps (Bönecke et al., 2021; Kuang et al., 2014; Lund et al., 2005; Vogel et al., 2022). Despite this, sensor-based VRL is rarely adopted by farmers (Erickson & Lowenberg-Deboer, 2022). Most arable fields are still limed uniformly or managed on larger zones.
A major challenge for the adoption of sensor-based VRL lies in the high initial costs of creating the soil maps (Lowenberg-DeBoer, 2019). Consequently, making VRL more attractive to farmers is a matter of reducing its cost. While measurements of proximal soil sensors are cheap compared to conventional wet chemistry soil analysis (Adamchuk et al., 2004), most sensors do not measure the target soil properties directly but deliver so-called covariates which are related to the target soil properties. Following the principles of digital soil mapping (DSM), the relationship between a soil property and covariates can be exploited via a prediction model (McBratney et al., 2003). Yet, the training of a prediction model requires a certain number of training soil samples that have to be analyzed in the laboratory. Since laboratory analyses are expensive, the size of the training sample set should be kept small. Nonetheless, the sample set should not be too small either, as this would result in poorly fitted prediction models (Lucà et al., 2017; Ramirez-Lopez et al., 2014). Currently, there is a lack of clear guidelines on how many training samples are ideally needed in the context of sensor-based PA. Therefore, in practice, the number of soil samples is often determined by what a client is willing to pay and not necessarily by what is appropriate.
The above indicates that further research is needed to determine adequate training sample sizes for sensor-based soil mapping. Logically, larger training sample sizes result in more accurate predictions but the relationship between the training sample size and model accuracy seems to be non-linear and tends to saturate with increasing sample size (Chen et al., 2021; Lucà et al., 2017; Ramirez-Lopez et al., 2014). The point at which the model accuracy plateaus and after which no meaningful model improvements are achieved depends on several factors, such as the spatial extent of the study area (Ramirez-Lopez et al., 2014), the spatial variability, the target soil property, the deployed prediction model or the explanatory power and the dimensionality of the available covariates (Sun et al., 2022). Therefore, the optimal sample size is case-dependent but more research could assist in finding adequate training sample sizes.
Besides the sample size, attention should also be paid to the sampling design and prediction model, as the right combination of these aspects may help increase the prediction accuracy while reducing the number of necessary training samples. Sampling designs have shown a considerable influence on prediction performances (Biswas & Zhang, 2018). In the context of DSM, it is usually desired that a sampling design spreads soil samples equally in the geographical space or that the whole range of values of the covariate space are covered (Brus, 2019; Brus et al., 2006). However, recent studies indicate that coverage of the covariate space is more important than geographical coverage when covariates with significant explanatory power are available (Biswas & Zhang, 2018; Wadoux et al., 2019). Several sampling designs have been developed for achieving an optimal spread in the covariate space but most commonly used are conditioned Latin hypercube sampling (cLHS) (Minasny & McBratney, 2006) and k-means sampling (KM) (Brus, 2019). In contrast, simple random sampling (SRS) is neither optimized to cover the geographical nor the covariate space but can function as a baseline performance.
Sampling designs are often not fully deterministic but have a random component in their sampling procedure. This means that when the sampling is repeated, it may not select the exact same sampling sites again. Consequently, the new underlying training data would generate different prediction models, leading to varying model performances. Wadoux and Brus (2021) showed that it is therefore crucial to repeat the sampling when comparing the performance of competing sampling designs. This approach enables the comparison of sampling designs based on summary measures of model performances across multiple repetitions. SRS is a fully randomized sampling design but also cLHS and to some extent KM have a random component in their sampling procedure. Some studies that did not repeat the sampling seemed to favor cLHS as being most effective for DSM (Castro-Franco et al., 2015; Schmidt et al., 2014) but according to Wadoux and Brus (2021) these may be incidental outcomes, as studies with a repetitive design contested the efficiency of cLHS compared to KM or even SRS (Ma et al., 2020; Wadoux & Brus, 2021; Wadoux et al., 2019). Finally, Ng et al. (2018) and Wang et al. (2021) also used a repetitive study design, in which both cLHS and KM proved to be effective. These conflicting results indicate that there is still much uncertainty about the effectiveness of different spatial sampling designs.
In the context of sensor-based VRL, primarily linear regression models have been utilized for predicting the required soil properties (Bönecke et al., 2021; Schirrmann et al., 2011; Vogel et al., 2022). Nonetheless, machine learning techniques like Random Forest (RF) could enhance the prediction accuracy, due to their ability to deal with non-linear relationships and interactions (Chen et al., 2022). However, such machine learning algorithms typically require larger training sample sizes to fit models that generalize well.
The main objective of this study was to assess which combination of spatial sampling design and prediction model yields sufficiently high prediction accuracies for mapping clay, SOC and pH in an agricultural field, while using as few soil samples as possible. For this, a repetitive sampling procedure was implemented to investigate the combined effects of: (i) training sample size; (ii) spatial sampling design; and (iii) prediction model.
Materials and methodology
Study area
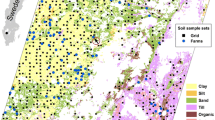
The test field is located in the Eastern part of Brandenburg, Germany (Fig. 1) and has a size of approximately 51 ha. The area was strongly influenced by recurring glacial deposition and aeolian processes during the peri- and interglaciation, which led to predominantly sandy soils. Nonetheless, on a smaller spatial scale, soils may show considerable heterogeneity due to diverse and alternating geomorphic processes (Vogel et al., 2022). The elevation of the test field ranges from 48 to 70 m a.s.l. The study area is located in the transitional zone between oceanic and continental climate, leading to rather dry conditions compared to the rest of Germany. It has an average annual rainfall of 550 mm, which is relatively evenly distributed throughout the year. The annual mean temperature is 9 °C, with January being the coldest month with a mean of 0 °C and July the warmest with a mean of 20 °C.
Reference soil samples
In May 2020, 250 reference soil samples were taken from the topsoil (0–30 cm) on a triangular pattern with 50 m distance to the nearest six neighbors. Each soil sample was composed of subsamples taken in a radius of 9 m around the center. The composite soil samples had a total weight of approximately 1 kg. These samples were oven-dried at 75 °C, sieved to < 2 mm and then analyzed in the laboratory on soil organic carbon (SOC) content (g g −1), clay content (g g −1) and pH value.
The pH value was measured in 10 g of soil and 25 ml of 0.01 M CaCl2 solution according to DIN ISO 10390 with a glass electrode after an equilibration time of 60 min. The particle size distribution of the < 2 mm fraction was determined according to DIN ISO 11277 by wet sieving and sedimentation after removal of organic matter with H2O2 and dispersion by 0.2 N Na4P2O7. SOC was analyzed by elementary analysis using the dry combustion method (DIN ISO 10694) after removing the inorganic carbon with hydrochloric acid. All the data used for this study will be freely available by the end of the I4S Project but can be granted on request before. Summary statistics can be found in Table 1.
Geostatistical simulation
A sampling design may select sample-sites at any point in the geographical domain of the study area. Yet, the soil attributes are only known at the 250 sample locations (Fig. 1), meaning that for the majority of the study area, the values of the soil attributes were unknown. Therefore, geostatistical simulations were used to stochastically simulate the unknown values and generate so-called ‘possible realities’. Each simulation functioned then as a distinct reference map. This permitted the repetition of the sampling selection to create multiple training sample sets from every reference map (“Experimental design” Section).
The simulations were generated with the sequential Gaussian simulation algorithm (Goovaerts, 1997). In this algorithm, given a semivariogram (Fig. 2) and a list of locations, the soil property is simulated sequentially for each location of the map based on a random draw from its conditional probability distribution. The conditional probability distribution is assumed to be normal distributed and derived from kriging and previously simulated values in the local neighborhood. Stationarity and isotropy were assumed, meaning that the geostatistical model had a constant mean and a semivariance that only depends on the Euclidean distance between locations. Note, that one may be tempted to use ordinary kriging to predict the expected value for each unknown location to generate only one single reference map instead of using multiple simulations. However, ordinary kriging would disregard the actual existing short-distance soil variation and would generate a reference soil map that is much smoother i.e., has much less short-distance variability, than the reality (Goovaerts, 1997). Using such a smoothed map as reference map in an experimental setting would ultimately result in overoptimistic performances. In contrast, with sequential Gaussian simulation, the spatial variability as assessed through the semivariogram is preserved. Yet, it requires that a set of multiple simulations is analyzed, as the unknown reality is treated as one of an infinite number of possible realities (Goovaerts, 1997; Heuvelink, 2018; Pebesma, 2004). A similar approach with geostatistical simulations can be found in Heuvelink et al. (2010).
For pH, SOC and clay, 100 sequential Gaussian simulations were created based on the 250 reference soil samples. The raster size was 8 × 8 m leading to an overall population of N = 8,035 locations per simulation. The simulations were executed using the package gstat (Pebesma, 2022) in the statistical language R (R Core Team, 2021a). Examples of these simulations for all three target soil properties can be seen in Fig. 3.
Covariates
The covariates were obtained by two mobile multi-sensor platforms: the Veris® Mobile Sensor Platform 3 (MSP3) (VERIS Technologies, Salinas, KS, USA) and the Geophilus platform (Lück & Rühlmann, 2013). The combination of these two multi-sensor platforms was established in previous projects within the context of VRL. We refer to Bönecke et al. (2021) for more technical background information about the sensors.
In August 2021, 998 measurements were taken along transects by Veris MSP3 and on the same day, 14,288 measurements were obtained by the Geophilus platform (Fig. 4). As can be seen in Fig. 4, some areas along the transects of the Veris MSP3 were undersampled due to measurement errors, such as when a sensor was blocked by stones or organic material.
The Veris MSP3 was mounted with multiple proximal soil sensors. This includes an optical soil sensor (OpticMapper) measuring the reflectance with a photodiode at 660 nm (red) and at 940 nm, and a soil pH manager using two ion-selective antimony electrodes to measure the pH value. The pH was measured on naturally moisturized soil and partly from tap water used to clean the antimony electrodes. The Veris MSP3 was additionally mounted with the capacitance sensor of the Veris iScan that was measuring the volumetric soil moisture (%) (Fig. 5a–c). Note that the pH measured by the pH manager is only a proxy of the standard lab-measured pH value, as different measuring devices are used and external environmental factors may confound the in-situ measurements (Vogel et al., 2023). Hence, the pH measurements obtained by the Veris MSP3 are usually not used as the direct target variable but serve as covariate (Lund et al., 2005). In the following, it is referred to as sensor pH, to distinguish it from the pH obtained from the reference soil samples.
The Geophilus platform consists of a multiple rolling electrodes array to measure apparent electrical resistivity (ER in Ω·m) of different soil depths and a gamma sensor, detecting total counts of gamma-ray activity of the topsoil. ER measurements were discarded when the frequency of the measured voltage deviated from the frequency of the inserted current by a set threshold of + / − 30 Hz. Additionally, the altitude (m a.s.l.) was taken from a differential global navigation satellite system (dGNSS) (Fig. 5d–f).
Summary statistics of the measurements of each sensor are given in Table 1. All produced covariates were interpolated using ordinary kriging to raster maps with 8 m × 8 m spatial resolution. Furthermore, the min. to max. kriging standard deviation (stdkrig) was given to indicate the uncertainty of the maps (Fig. 5).
Measurements of soil reflectance at 940 nm and soil electrical conductivity by Veris MSP3 were not included in the final covariate space for the analysis. These were removed in the pre-processing because they had a large correlation with red and ER, respectively. The decision to drop these covariates was based on what we expected to have greater explanatory power, with knowledge from prior studies within the same project (e.g. Bönecke et al., 2021; Vogel et al., 2022). Not accounting for multicollinearity might hamper the effectiveness of sampling designs that use the covariate space because it gives less weight to covariates that have no or less correlation with other covariates (Brus, 2019).
Experimental design
The workflow of the study is given in Fig. 6. The sensor-based covariate maps and the 100 sequential Gaussian simulations of clay, SOC and pH were used as inputs. Three sampling designs, SRS, cLHS and KM, repeatedly selected samples from the simulated reference maps to create multiple training sample sets of different sample sizes for each individual simulated reference map. The sample sizes ranged from 5 to 30, with small sample size increments and from 30 to 100 with larger increments. Smaller sample sizes were overrepresented because usually smaller sample sizes are used in practical agriculture due to financial constraints (e.g. Adamchuk et al., 2011; Bönecke et al., 2021). Furthermore, the used proximal soil sensor combinations (covariates) were optimized on the target soil properties (Bönecke et al., 2021).
The sample selection used to generate the training sample sets was repeated 150 times for each sample size, sampling design and simulation. Every training sample set was then used to fit random forest (RF) and multiple linear regression (MLR) models for pH, clay and SOC predictions. This led to a total number of 900,000 individual prediction maps for each target soil property (10 training sample sizes × 3 sampling designs × 2 prediction models × 150 sampling repetitions × 100 simulations).
Finally, the results were compared to a uniform management approach, where the mean value of the field is assumed to be representative of the entire field. The average value of pH, clay and SOC was calculated from all 100 aggregated Gaussian simulations. With this approach, the within-field variability is not taken into account. Nonetheless, the uniform management approach is still commonly used in practice in which the mean value is approximated based on a composite sample.
Since each Gaussian simulation is treated as one simulated reality of the target soil property, the entire map can be used for validation. Thus, the prediction map is validated by comparing it to the simulation. Normally, it is not advised to use training samples for validation (Piikki et al., 2021) but since the used training sample sets are only a very small subsample compared to the overall population (n < < N; n ≤ 100, N = 8,035), this should not lead to overoptimistic results. As validation metrics, the root-mean-square error (RMSE), mean error (ME) and Lin’s concordance correlation coefficient (CCC) was used.
Sampling designs
SRS is the most straightforward sampling design and does not require any prior knowledge about the study area nor expertise in its deployment. Each soil sampling location is drawn independently from the area and all locations have equal probability to be chosen. The performance of SRS can be used as a baseline when compared with the performance of the other two purposive sampling designs, because if a sampling design is not better than SRS, there would be no justification to use it.
cLHS (Minasny & McBratney, 2006) is an adaptation of the classic Latin hypercube sampling algorithm, specifically designed for soil mapping studies. It stratifies the covariate space by dividing the domain of each of the c covariates into n equally sized marginal strata based on its cumulative distribution, where n is also the sample size. The goal is to cover each stratum for every covariate with one sample and simultaneously preserve the correlation between covariates similar to that of the original covariate space. This is done by minimizing the weighted sum of two objective functions:
where zij is the number of samples in stratum i for covariate j. Further, apq and bpq are elements of a correlation matrix from the original data and the sampled sites, respectively, and are determined for each covariate combination p and q. cLHS was executed using the clhs R package (Roudier, 2021), in which the objective function is minimized through a heuristic search algorithm based on simulated annealing. Default weight values (w1 = w2 = 1) and 10,000 iterations were used. As simulated annealing is a probabilistic numerical optimization algorithm, it does not always come up with the same and best solution i.e., the global optimum (Bertsimas & Tsitsiklis, 1993). Therefore, O and the associated selected sample sites are not fixed, leading to a random component within cLHS.
In KM, the dataset is stratified by building k clusters using the k-means algorithm, where k is equal to the sample size (Brus, 2019). The clusters are built based on the similarity of the data in the covariate space. This means that a data point belongs to the cluster whose centroid is the nearest in terms of the Euclidean distance. The initial centroids are set randomly but they are repeatedly recomputed and adjusted until the squared sum of distances between the data points and the cluster centroids are minimized as given by the objective function J:
where δij is a binary variable that is 1 if sample vi belongs to a cluster and 0 if not. Moreover, dj is the centroid of the cluster. The k-means algorithm may also not always be able to determine the global optimum i.e., the condition with the lowest value for J, as the final clusters depend on the random selections at the initialization phase. The risk to end up in a local optimum can be minimized by restarting the clustering several times with different initial clusters and taking the outcome with the lowest value of the objective function (Hartigan & Wong, 1979; Steinley, 2003). In this study, KM returned the same or very few different sample configurations, meaning that it most likely often found the global optimum. However, this was not necessarily desired in our study design, as this would mean that the performance of KM would be judged based on one or few sample configurations. Therefore, two sources of randomization were artificially introduced. Firstly, the older Lloyd version (Lloyd, 1982) was used, in which the random initial clustering is not repeated. Secondly, KM was modified by not taking the centroid as the representative sample itself but by randomly selecting one of the nine nearest samples (in terms of covariate space) or the centroid. The clustering of the data was conducted via the kmeans function of the stats R package (R Core Team, 2021b) and with standardized covariates.
Prediction models
Machine learning techniques have manifested themselves as state-of-the-art for DSM modeling, as they are able to deal with non-linear relationships and untangle complex patterns within the data (Chen et al., 2022). RF is a tree-based machine learning technique, in which through averaging of an ensemble of regression trees a prediction value is derived (Breiman, 2001). It showed advantages over conventional spatial models (Hengl et al., 2018). It is often not recommended to use data-driven machine learning models when only very few training data are available, as may often be the case in practical PA. Nonetheless, RF was used in this study due to the fact that Schmidt et al. (2014) and Ma et al. (2020) obtained satisfactory prediction accuracy with RF for a small sample size of 20. Furthermore, the goal of this study was also to observe the limitations of the methods at very small sample sizes. RF modeling was done with the randomForest R package (Liaw & Wiener, 2022).
Despite the popularity of machine learning techniques, linear models are still in many cases used for DSM (Chen et al., 2022). Almost all studies in the context of sensor-based VRL used linear models (Bönecke et al., 2021; Lund et al., 2005; Vogel et al., 2022). For this study, covariates were selected once beforehand based on the original dataset. This was done through a backward elimination with a significance threshold of 0.025 for the t-test but no sample size correction was applied to account for spatial autocorrelation. SOC and ER were log-transformed prior to fitting a MLR to linearize non-linear relationships and better meet the assumption of residuals being normally distributed. This resulted in clay being predicted by log(ER), gamma and sensor pH, SOC being predicted by log(ER), sensor pH, altitude and red, and lastly pH being predicted by sensor pH and log(ER). Before calculating the validation metrics, the predicted log(SOC) values were back-transformed to express the RMSE and ME in terms of SOC. A ‘smearing’ retransformation (Duan, 1983) was used to correct for the bias that would be incurred by a ‘naive’ back-transform based on the inverse of the logarithm.
Validation metrics
Root-mean-square error (RMSE) is the most commonly used validation metrics in the context of DSM (Piikki et al., 2021). It indicates the deviation between the predicted and validation value i.e., the prediction error. Thereby, it considers both systematic and random prediction errors and is non-negative:
where \(\widehat{y}\)(li) and y(li) are the predicted and observed soil property at location li, respectively.
As mentioned before, a larger training sample size is expected to improve model performance. To measure the relative improvement, the information gain (IG) was introduced. IG indicates the relative improvement of the median RMSE in percent over all geostatistical simulations and iterations that can be obtained with one extra soil sample. It is calculated by the difference between a larger sample size set compared to the previous sample size set divided by the difference of soil samples.
The mean error (ME) indicates the systematic error (Piikki et al., 2021) and can have negative and positive values:
The RMSE and ME cannot be used to compare the prediction performance of the different soil properties because they present the error in the associated measurement unit. Hence, a dimensionless, scaled performance index is required. Several of these indices are available (Piikki et al., 2021). Among them, the concordance correlation coefficient (CCC) (Lin, 1989) offers the advantage to account for random as well as systematic errors:
where ρ is the Pearson correlation coefficient between the predicted and reference value. Correspondingly, \(\widehat{s}\) 2 (predicted) and s2 (reference) are the variances and \(\widehat{\overline{x}}\) (predicted) and \(\overline{x}\) (reference) are the mean values. Therefore, bias and precision are considered simultaneously. CCC ranges from -1 to 1, where a higher value represents better agreement. Viscarra Rossel et al. (2016) defined as approximate benchmarks that a CCC < = 0.65 denotes poor agreement, 0.80 > = CCC > 0.65 moderate agreement, 0.90 > = CCC > 0.80 substantial agreement and CCC > 0.90 excellent agreement.
Results
Effect of sample size
For all three target soil properties, RMSE and CCC strongly depended on the sample size. As expected, larger sample sizes led to a decrease in the median RMSE (Fig. 7a–c, Tables 2, 3) and an increase in the median CCC (Fig. 7 d–f, Table 4). Simultaneously, the SD of the RMSE distribution decreased with increasing sample size (Fig. 8, Tables 2, 3). The median RMSE decreased between sample size 5 to 100 by 29% for clay, 53% for SOC and 27% for pH. The RMSE values with 100 samples were 1.73 for clay, 0.21 for SOC and 0.3 for pH.
Despite this constant improvement, the relative improvement, as seen from the IG, decreased with increasing sample size (Figs. 7, 8, Tables 2, 3). The IGs from sample size 5 to 10 ranged from 0.57% to 6.23% for clay, 3.29% to 12.94% for SOC and 0.92% to 4.3% for pH. In contrast, the maximum IGs achieved with 75 to 100 soil samples were 0.071%, 0.13% and 0.1% for clay, SOC and pH, respectively. This is further reflected in the flattening, i.e. saturation, of the curve in Fig. 7.
For all three target soil properties, the sensor-based predictions outperformed the uniform estimations by far with at least 10 training samples (Fig. 7). When using only 30 training samples, the median RMSE was in comparison to the uniform approach approximately 20%, 50% and 25% lower for clay, SOC and pH, respectively.
The ME, given in Table 5 in the Appendix, indicated bias in some circumstances for small sample sizes between 5 and 10. However, with increasing sample size ME approached zero in most cases.
Effect of prediction model
The saturation of the model improvement, seen from the flattening of the graph in Fig. 7, was more pronounced for MLR models than for RF models. After some point, MLR plateaued completely and the sample size did not noticeably affect model performance anymore. On the other hand, RF models significantly benefited from additional soil samples even at larger sample sizes. This behavior is especially characterized by the IG (Table 2, 3). Between sample size 75 to 100, IG for MLR was between 0.015% to 0.035%, whereas for RF it was between 0.063% to 0.128%. Yet, for MLR, values below 0.1% were already achieved at sample size 50, 75 and 40 for clay, SOC and pH, respectively. Consequently, RF continuously improved its performance substantially and did not plateau completely. At smaller sample sizes, the results were less straightforward and dependent on the soil property. Judging from the median RMSE, in case of clay, RF performed best regardless of the sample size. However, for SOC and especially for pH, lower median RMSE values were obtained with MLR. Nonetheless, RF outperformed MLR at a sample size of 40 for SOC and 75 for pH. When looking at the mean RMSE, RF performed better than MLR at sample size 5 for all soil properties (Tables 6, 7). The small mismatch at sample size 5 between median and mean RMSE is due to the high SD of MLR models at this sample size (Tables 6, 7). With increasing sample size, the SD of MLR decreased rapidly, so that it eventually was similar for MLR and RF.
Furthermore, we observed a small mismatch between median CCC and median RMSE results (Tables 2, 3, 4). Especially at smaller sample sizes (5 to 40), CCC tended to be higher i.e. better for MLR compared to RF. In the case of clay, CCC was larger for MLR than RF, while RMSE showed the opposite. Hence, around 30 to 40 samples were needed before RF scored better CCC values than MLR.
In general, CCC values already indicated moderate agreement (CCC > 0.65) at a small sample size of around 10 for all soil properties with most combinations (Fig. 7d–f, Table 4). However, neither for clay nor pH, models of substantially high quality (CCC > 0.8) were achieved regardless of the sample size, whereas excellent agreement (CCC > 0.9) was found for SOC when using 100 samples and RF.
ME values (Table 5), deviated more from zero for MLR at sample size 5. However, with increasing sample size ME was rather unaffected by the prediction model.
Effect of sampling design
The RMSE of the prediction models varied between the sampling designs applied. Generally, SRS received a higher RMSE and lower CCC value compared to cLHS and KM across almost all sample sizes and target soil properties (Figs. 7, 8, Tables 2, 3, 4). This difference was especially manifested at smaller sample sizes, where SRS performed very poorly. At larger sample sizes, the difference decreased and in the case of MLR, the performance of all three sampling designs was nearly the same once the plateau of the graph was reached. For example, at sample size 5 the median RMSE of SRS was around 17.4% larger than of cLHS and KM, whereas at sample size 100 it was only 0.8% larger.
Based on all sample sizes and soil properties, neither KM nor cLHS clearly outperformed each other, although dissimilarities in their performance can be seen. However, these differences seemed to have no general pattern. The only visual trend was that KM seemed to perform better at larger sample sizes but was less coherent and stable at smaller sample sizes (Figs. 7, 8). On a few occasions such as for SOC at sample size 20 to 25, this led to even poorer performances than SRS. In general, the difference between KM and cLHS was low.
There was no clear dependency between the sampling design and ME (Table 5) apart from KM for clay. In this case, slightly positive biases were found for both RF and MLR.
Discussion
Effect of sample size and prediction model
As expected, the accuracy of the predictions was largely determined by the training sample size and to a certain smaller degree by the prediction model. It can generally be inferred that ‘the larger the training sample size the better the prediction’, although the relative improvement decreased as the sample size got bigger. Furthermore, MLR tended to perform better at lower sample sizes, while RF was more powerful at larger sample sizes. However, the best model performance also depended strongly on the target soil property and the validation metric considered. In some cases, RMSE favored RF over MLR while the opposite was found for CCC. A similar discrepancy was reported in Somarathna et al. (2017) for several machine learning models, including RF. At sample size 5, ME values indicated considerable bias. This is likely due to a few extreme values (Fig. 8) because after a sample size of 10 and larger, bias mostly disappeared.
Judging from both RMSE and CCC, a sample size of around 30 to 40 was needed before RF outperformed MLR for clay and SOC. This is unsurprising as RF is able to deal with non-linear relationships and interactions (Chen et al., 2022) but it simultaneously relies on more data to fit a richer model. For pH, the outcome was different. MLR clearly outperformed RF. One reason may be that fewer covariates were considered in the fitting of the MLR model. Further, it is known from prior studies that the sensor output of the ion-selective pH electrodes (sensor pH) used in the Veris MSP3 has a strong linear correlation with the standard laboratory pH value (Bönecke et al., 2021; Lund et al., 2005; Schirrmann et al., 2011). Hence, MLR was much more powerful at small sample sizes compared to RF. However, with at least 75 samples, RF also outperformed MLR for pH. Yet, it has to be noted that we did a variable selection beforehand with the original reference dataset, which is impossible in practice. If all covariates would be considered simultaneously in an MLR, most likely worse results would be found at lower sample sizes. Therefore, there is a need to define relevant variables for MLR from multiple campaigns to come up with robust mechanistic models.
The advantage of machine learning techniques like RF over linear models is not surprising when a sufficient number of data points is available for training (Hengl et al., 2018). However, our results show that RF can also perform well at relatively small sample sizes, where the use of machine learning models would normally not be recommended. Our findings are in accordance with those reported in Schmidt et al. (2014) and Ma et al. (2020), which also observed an adequate fit with RF compared to other models, when using only 20 soil samples.
The prediction models may be further improved by adding a spatial component such as in regression kriging (Hengl et al., 2004). However, this requires a large dataset to fit a residual variogram (Webster & Oliver, 2007) and the added value might be small because of weak residual spatial autocorrelation (Heuvelink & Webster, 2022).
Effect of sampling design
The sampling design significantly affected the accuracy of the prediction models. Both KM and cLHS proved to be beneficial for modeling, due to their superiority over SRS. This confirms the importance of having a wide coverage of the covariate space. From the results, it is not possible to either favor KM or cLHS. There was only a minor apparent trend that corresponds to findings in Ng et al. (2018) and Wang et al. (2021): KM was more likely to perform better at larger sample sizes but cLHS was generally robust over all sample sizes. Nonetheless, the fluctuating behavior of KM at lower sample sizes might be explained by its more deterministic sample selection. This led to less variation between the selected training samples to build up its training sample sets. Therefore, the models relied more or less on the same training samples in the fitting process at specific training sample sizes. In some cases, these may by accident, have been less optimal. Moreover, this likely explains why a slightly positive bias can be found for clay when using KM. A similar behavior of KM can be found in Wadoux et al. (2019). By slightly adapting the KM to introduce randomization (“Sampling designs” Section), it was attempted to counteract this behavior but it could not be eliminated entirely. Note, that the modification may also have affected the prediction performance to some unknown extent.
The strong performance of cLHS was surprising because a similar methodology and recommendations from Wadoux et al. (2019), Ma et al. (2020) and Wadoux and Brus (2021) were adopted but fundamentally different results with regards to cLHS were obtained. In those studies, cLHS did not perform significantly better than SRS, suggesting that other sampling designs such as KM should be preferred within DSM. Our results do not fully support this conclusion. At no sample size was the average performance of cLHS worse than that of SRS. More recently, other studies showed similar results (Brus, 2022; Wang et al., 2022), further strengthening that cLHS may indeed be a useful sampling design for soil mapping.
When comparing other case studies, sampling designs performed inconsistently, even though similar methodologies were used (Brus, 2022; Ma et al., 2020; Ng et al., 2018; Wadoux & Brus, 2021; Wadoux et al., 2019; Wang et al., 2021). Hence, more research about the reasons that determine the success of a sampling design is needed. Additionally, modifications of KM and cLHS algorithms may be tested to further improve performances. This might include different weights for the optimization criterion in cLHS.
Implications for precision liming
The use of sensor-based DSM resulted in far better predictions compared to the still commonly used uniform approach (Fig. 7). A relatively small number of soil samples (> 10) were sufficient to outperform the uniform approach. With 20–30 soil samples the difference was already very pronounced. This underlines the effectiveness and potential benefit of deploying sensor-based soil mapping for VRL.
The RMSE curve of MLR leveled off at medium training sample sizes, after which no significant further model improvements were achieved. Hence, RF generated the best prediction models with a medium to large sized training sample set. Since the RF curve did not completely level it appears that RF models would still improve at even larger sample sizes than evaluated in this study. Given that no economic analysis was included, it is not possible to evaluate to which point the model improvement justifies the associated extra sampling costs. While more training samples always proved to be better, the sharp decrease in relative model improvement makes it contestable if very large sample sizes are necessary. It appears that the improvement for sample sizes > 40 is probably not large enough to have significance on liming recommendations and thus yield, when considering the extra sampling costs and low IG. Bönecke et al. (2021) raised the question if it is possible to reduce the number of training samples per field to a maximum of 5 to 10 for the same proximal soil sensor combination in order to reduce laboratory costs. Figure 8 shows that reasonable results can be obtained in some cases at such a small sample size but the risk to receive an insufficient prediction model is very high due to the high standard deviation of the RMSE for very small sample sizes. Even though not considered in this study, a larger sample size is also advisable to allow validation (e.g. leave-one-out cross-validation) to verify the accuracy of the predicted soil maps. Therefore, using only a few samples (< 10) in order to cut costs should be avoided. Only a few extra soil samples would already substantially increase model accuracy and robustness. Nonetheless, for our study field, relatively stable results for SOC and clay were obtained with 20 to 40 soil samples and even fewer than 20 for pH.
Limitations and future research
For future studies on sample size optimization in the context of sensor-based PA, it might be beneficial to include an economic analysis. In order to come up with an optimal sample size, the sampling costs have to be weighed against the expected yield loss resulting from incorrect liming recommendations caused by prediction errors (Lowenberg-DeBoer, 2019). Moreover, the use of proximal soil sensors per se is already associated with higher costs.
It has to be emphasized that the present results are based on a single case study and restricted in their validity to the specific sensor combination used in this study. When using other sensors, very different outcomes may be observed. One would expect that more training data are needed for accurate prediction when the covariate space has a large dimensionality. For example, other studies, in which multiple bands within the whole visible and near-infrared spectrum were used, indicate that much larger training sample sizes are desirable (Debaene et al., 2014; Lucà et al., 2017). Additionally, the effect of spatial factors such as field size and soil variability cannot be considered within a single field. It is expected that a larger sample size is needed for bigger fields, because bigger fields tend to have more soil variation (Chen et al., 2021; Ng et al., 2018).
Since a geostatistical simulation was used instead of a complete dataset with real values, our final results also depend on the assumed validity of the geostatistical model. Hence, the absolute values of RMSE and CCC have to be interpreted with care (Ma et al., 2020; Wadoux & Brus, 2021). Yet, the RMSE of e.g., pH is similar with results reported in the literature in which an RMSE of 0.28 to 0.55 (Lund et al., 2005) or 0.37 (Bönecke et al., 2021) was found when using the Veris MSP3 at training sample sizes of around 30. The study indicates that the use of geostatistical simulations for sampling designs are a reasonable alternative for small scale study areas with moderately sized datasets. However, the implications of using geostatistical simulations based on a real dataset compared to using the dataset directly as in Ng et al. (2018), Wadoux et al. (2019) or Wadoux and Brus (2021) could be subject of further research.
Conclusion
By assessing the impact of training sample size, spatial sampling design and prediction model on sensor-based soil mapping of clay, SOC and pH, it was observed that while increasing the training sample sizes improved the prediction accuracy, the relative improvement decreased continuously. The plateauing improvement in accuracy was more pronounced with MLR compared to RF. Hence, RF performed considerably better at larger sample sizes. Yet, MLR showed advantages at smaller sample sizes. The results suggest that a sample size ranging between 10 to 40 training samples appears reasonable for the employed sensor combination. With MLR, prediction models of moderate quality (CCC > 0.65) were already fitted using only 10 training samples for all three target soil properties. Nonetheless, 10 training samples are the lowest boundary, as slightly more training data ensures better model quality and stability. At smaller training sample sizes (< 10), the probability to receive insufficient prediction models is very high due to the large standard deviation of the RMSE. Models of excellent quality (CCC > 0.9) were only obtained for SOC at a sample size of 100 with RF. cLHS and KM proved to be effective sampling designs compared to SRS but neither cLHS nor KM clearly outperformed each other. In contrast to other studies, cLHS performed better than SRS. While a sampling design and prediction model can enhance the prediction accuracy, having a larger sample size proved to be the most crucial factor for accuracy at the low to medium sample size range. Sensor-based soil mapping yielded much better results compared to a uniform approach, almost regardless of sample size, sampling design and prediction model.
Note that these recommendations are derived from a single case study and restricted to the used sensor combination. Nonetheless, the results may help improve current practices in sensor-based precision liming or site-specific soil management in general. For more general conclusions, more studies are needed from different soilscapes and fields of different soil variability or size.
References
Adamchuk, V. I., Morgan, M. T., & Lowenberg-Deboer, J. M. (2004). A model for agro-economic analysis of soil pH mapping. Precision Agriculture, 5, 111–129. https://doi.org/10.1023/B:PRAG.0000022357.28154.eb
Adamchuk, V. I., Viscarra Rossel, R. A., Marx, D. B., & Samal, A. K. (2011). Using targeted sampling to process multivariate soil sensing data. Geoderma, 163, 63–73. https://doi.org/10.1016/j.geoderma.2011.04.004
Bertsimas, D., & Tsitsiklis, J. (1993). Simulated annealing. Statistical Science, 8(4), 10–15. https://doi.org/10.1214/ss/1177011077
Biswas, A., & Zhang, Y. (2018). Sampling designs for validating digital soil maps: A review. Pedosphere, 28, 1–15. https://doi.org/10.1016/S1002-0160(18)60001-3
Bönecke, E., Meyer, S., Vogel, S., Schröter, I., Gebbers, R., Kling, C., Kramer, E., Lück, K., Nagel, A., Philipp, G., Gerlach, F., Palme, S., Scheibe, D., Zieger, K., & Rühlmann, J. (2021). Guidelines for precise lime management based on high-resolution soil pH, texture and SOM maps generated from proximal soil sensing data. Precision Agriculture, 22, 493–523. https://doi.org/10.1007/s11119-020-09766-8
Breiman, L. (2001). Random forests. Machine Learning, 45, 5–32. https://doi.org/10.1023/A:1010933404324
Brus, D. J. (2019). Sampling for digital soil mapping: A tutorial supported by R scripts. Geoderma, 338, 464–480. https://doi.org/10.1016/j.geoderma.2018.07.036
Brus, D. J. (2022). Spatial sampling with R. CRC Press.
Brus, D. J., de Gruijter, J. J., & van Groenigen, J. W. (2006). Designing spatial coverage samples using the k-means clustering algorithm. Developments in Soil Science, 31, 183–192. https://doi.org/10.1016/S0166-2481(06)31014-8
Castro-Franco, M., Costa, J. L., Peralta, N., & Aparicio, V. (2015). Prediction of soil properties at farm scale using a model-based soil sampling scheme and random forest. Soil Science, 180, 74–85. https://doi.org/10.1097/SS.0000000000000115
Chen, S., Arrouays, D., Mulder, V. L., Poggio, L., Minasny, B., Roudier, P., Libohova, Z., Lagacherie, P., Shi, Z., Hannam, J., Meersmans, J., Richer-de-Forges, C. A., & Walter, C. (2022). Digital mapping of GlobalSoilMap soil properties at a broad scale: A review. Geoderma, 409, 115567. https://doi.org/10.1016/j.geoderma.2021.115567
Chen, S., Xu, H., Xu, D., Ji, W., Li, S., Yang, M., Hu, B., Zhou, Y., Wang, N., Arrouays, D., & Shi, Z. (2021). Evaluating validation strategies on the performance of soil property prediction from regional to continental spectral data. Geoderma, 400, 115159. https://doi.org/10.1016/j.geoderma.2021.115159
Debaene, G., Niedźwiecki, J., Pecio, A., & Żurek, A. (2014). Effect of the number of calibration samples on the prediction of several soil properties at the farm-scale. Geoderma, 214, 114–125. https://doi.org/10.1016/j.geoderma.2013.09.022
Duan, N. (1983). Smearing estimate: A nonparametric retransformation method. Journal of the American Statistical Association, 78, 605–610. https://doi.org/10.2307/2288126
Erickson, B., & Lowenberg-Deboer, J. M. (2022). 2022 Precision agriculture dealership survey. Purdue University. Retrieved January 30, 2023, from https://ag.purdue.edu/digitalag/_media/croplife-report-2022.pdf
Gebbers, R., & Adamchuk, V. I. (2010). Precision agriculture and food security. Science, 327, 828–831. https://doi.org/10.1126/science.1183899
Goovaerts, P. (1997). Geostatistics for natural resources evaluation. Oxford University Press.
Hartigan, J. A., & Wong, M. A. (1979). Algorithm AS 136: A k-means clustering algorithm. Applied Statistics, 28, 100. https://doi.org/10.2307/2346830
Hengl, T., Heuvelink, G. B. M., & Stein, A. (2004). A generic framework for spatial prediction of soil variables based on regression-kriging. Geoderma, 120, 75–93. https://doi.org/10.1016/j.geoderma.2003.08.018
Hengl, T., Nussbaum, M., Wright, M. N., Heuvelink, G. B. M., & Gräler, B. (2018). Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ, 6, e5518. https://doi.org/10.7717/peerj.5518
Heuvelink, G. B. M. (2018). Uncertainty and Uncertainty Propagation in Soil Mapping and Modelling. In A. McBratney, B. Minasny, & U. Stockmann (Eds.), Pedometrics. Springer.
Heuvelink, G. B. M., Jiang, Z., De Bruin, S., & Twenhöfel, C. J. (2010). Optimization of mobile radioactivity monitoring networks. International Journal of Geographical Information Science, 24(3), 365–382. https://doi.org/10.1080/13658810802646687
Heuvelink, G. B. M., & Webster, R. (2022). Spatial statistics and soil mapping: A blossoming partnership under pressure. Spatial Statistics, 50, 100639. https://doi.org/10.1016/j.spasta.2022.100639
Holland, J. E., Bennett, A. E., Newton, A. C., White, P. J., McKenzie, B. M., George, T. S., Pakeman, R. J., Bailey, J. S., Fornara, D. A., & Hayes, R. C. (2018). Liming impacts on soils, crops and biodiversity in the UK: A review. The Science of the Total Environment, 610–611, 316–332. https://doi.org/10.1016/j.scitotenv.2017.08.020
Kuang, B., Tekin, Y., Toby, W. W., & Mouazen, A. M. (2014). Variable rate lime application based on on-line visible and near infrared (vis-NIR) spectroscopy measurement of soil properties in a Danish field. Conference: Proceedings International Conference of Agricultural Engineering, Zurich, 06 - 10.07.2014.
Liaw, A., & Wiener, M. (2022). randomForest: Breiman and Cutler’s random forests for classification and regression. R Package Version, 4, 7–1.
Lin, L.I.-K. (1989). A concordance correlation coefficient to evaluate reproducibility. Biometrics, 45, 255. https://doi.org/10.2307/2532051
Lloyd, S. (1982). Least squares quantization in PCM. IEEE Transactions on Information Theory, 28(2), 129–137. https://doi.org/10.1109/TIT.1982.1056489
Lowenberg-DeBoer, J. (2019). The economics of precision agriculture. In J. Stafford (Ed.), Precision agriculture for sustainability. Burleigh Dodds Science Publishing.
Lucà, F., Conforti, M., Castrignanò, A., Matteucci, G., & Buttafuoco, G. (2017). Effect of calibration set size on prediction at local scale of soil carbon by Vis-NIR spectroscopy. Geoderma, 288, 175–183. https://doi.org/10.1016/j.geoderma.2016.11.015
Lück, E., & Rühlmann, J. (2013). Resistivity mapping with GEOPHILUS ELECTRICUS—Information about lateral and vertical soil heterogeneity. Geoderma, 199, 2–11. https://doi.org/10.1016/j.geoderma.2012.11.009
Lund, E., Adamchuk, V., Collings, K., Drummond, P., & Christy, C. (2005). Development of soil pH and lime requirement maps using on-the-go soil sensors. Precision Agriculture, 5, 457.
Ma, T., Brus, D. J., Zhu, A.-X., Zhang, L., & Scholten, T. (2020). Comparison of conditioned Latin hypercube and feature space coverage sampling for predicting soil classes using simulation from soil maps. Geoderma, 370, 114366. https://doi.org/10.1016/j.geoderma.2020.114366
McBratney, A., Mendonça Santos, M., & Minasny, B. (2003). On digital soil mapping. Geoderma, 117, 3–52. https://doi.org/10.1016/S0016-7061(03)00223-4
Minasny, B., & McBratney, A. B. (2006). A conditioned Latin hypercube method for sampling in the presence of ancillary information. Computers Geosciences, 32, 1378–1388. https://doi.org/10.1016/j.cageo.2005.12.009
Ng, W., Minasny, B., Malone, B., & Filippi, P. (2018). In search of an optimum sampling algorithm for prediction of soil properties from infrared spectra. PeerJ, 6, e5722. https://doi.org/10.7717/peerj.5722
Pebesma, E. (2022). Gstat: Spatial and Spatio-Temporal Geostatistical Modelling. Prediction and Simulation R package version 2. 0-9
Pebesma, E. J. (2004). Multivariable geostatistics in S: The gstat package. Computers Geosciences, 30, 683–691. https://doi.org/10.1016/j.cageo.2004.03.012
Piikki, K., Wetterlind, J., Söderström, M., & Stenberg, B. (2021). Perspectives on validation in digital soil mapping of continuous attributes—A review. Soil Use and Management, 37, 7–21. https://doi.org/10.1111/sum.12694
R Core Team 2021a R: A language and environment for statistical computing R Foundation for statistical computing
R Core Team. (2021b). stats: The R Stats Package. R package version 4.1.2 .
Ramirez-Lopez, L., Schmidt, K., Behrens, T., van Wesemael, B., Demattê, J. A., & Scholten, T. (2014). Sampling optimal calibration sets in soil infrared spectroscopy. Geoderma, 226–227, 140–150. https://doi.org/10.1016/j.geoderma.2014.02.002
Roudier, P. (2021). clhs: Conditioned latin hypercube sampling. R package version 0.7.0
Rühlmann, J., Bönecke, E., & Meyer, S. (2021). Predicting the lime demand of arable soils from pH value, soil texture and soil organic matter content. Agronomy, 11, 785. https://doi.org/10.3390/agronomy11040785
Schirrmann, M., Gebbers, R., Kramer, E., & Seidel, J. (2011). Soil pH mapping with an on-the-go sensor. Sensors, 11, 573–598. https://doi.org/10.3390/s110100573
Schmidt, K., Behrens, T., Daumann, J., Ramirez-Lopez, L., Werban, U., Dietrich, P., & Scholten, T. (2014). A comparison of calibration sampling schemes at the field scale. Geoderma, 232–234, 243–256. https://doi.org/10.1016/j.geoderma.2014.05.013
Söderström, M., Sohlenius, G., Rodhe, L., & Piikki, K. (2016). Adaptation of regional digital soil mapping for precision agriculture. Precision Agriculture, 17, 588–607. https://doi.org/10.1007/s11119-016-9439-8
Somarathna, P., Minasny, B., & Malone, B. P. (2017). More data or a better model? Figuring out what matters most for the spatial prediction of soil carbon. Soil Science Society of America Journal, 81, 1413–1426. https://doi.org/10.2136/sssaj2016.11.0376
Steinley, D. (2003). Local optima in k-means clustering: What you don’t know may hurt you. Psychological Methods, 8, 294–304. https://doi.org/10.1037/1082-989x.8.3.294
Stoorvogel, J., Kooistra, L., & Bouma, J. (2015). Managing soil variability at different spatial scales as a basis for precision agriculture. In R. Lal, B. A. Stewart, R. Lal, & B. A. Stewart (Eds.), Soil-specific farming: Precision agriculture. CRC Press.
Sun, X.-L., Lai, Y.-Q., Ding, X., Wu, Y.-J., Wang, H.-L., & Wu, C. (2022). Variability of soil mapping accuracy with sample sizes, modelling methods and landform types in a regional case study. CATENA, 213, 106217. https://doi.org/10.1016/j.catena.2022.106217
Viscarra Rossel, R. A., Behrens, T., Ben-Dor, E., Brown, D. J., Demattê, J., Shepherd, K. D., Shi, Z., Stenberg, H., Stevensi, A., Adamchuk, V., Aïchi, H., Barthès, B. G., Bartholomeus, H. M., Bayer, A. D., Bernoux, M., Böttcher, K., Brodský, L., Du, C. W., Chappell, A., … Ji, W. (2016). A global spectral library to characterize the world’s soil. Earth-Science Reviews, 155, 198–230. https://doi.org/10.1016/j.earscirev.2016.01.012
Vogel, S., Bönecke, E., Kling, C., Kramer, E., Lück, K., Philipp, G., et al. (2022). Direct prediction of site-specific lime requirement of arable fields using the base neutralizing capacity and a multi-sensor platform for on-the-go soil mapping. Precision Agriculture, 23, 127–149. https://doi.org/10.1007/s11119-021-09830-x
Vogel, S., Emmerich, K., Schröter, I., Bönecke, E., Schwanghart, W., Rühlmann, J., Kramer, E., & Gebbers, R. (2023). The effect of soil moisture content and soil texture on fast in situ pH measurements with two types of robust ion-selective electrodes. Egusphere. https://doi.org/10.5194/egusphere-2023-1470
Wadoux, A.M.-C., & Brus, D. J. (2021). How to compare sampling designs for mapping? European Journal of Soil Science, 72, 35–46. https://doi.org/10.1111/ejss.12962
Wadoux, A.M.-C., Brus, D. J., & Heuvelink, G. B. M. (2019). Sampling design optimization for soil mapping with random forest. Geoderma, 355, 113913. https://doi.org/10.1016/j.geoderma.2019.113913
Wang, J., Zhao, X., Zhao, D., & Triantafilis, J. (2021). Selecting optimal calibration samples using proximal sensing EM induction and γ-ray spectrometry data: An application to managing lime and magnesium in sugarcane growing soil. Journal of Environmental Management, 296, 113357. https://doi.org/10.1016/j.jenvman.2021.113357
Wang, Y., Qi, Q., Bao, Z., Wu, L., Geng, Q., & Wang, J. (2022). A novel sampling design considering the local heterogeneity of soil for farm field-level mapping with multiple soil properties. Precision Agriculture. https://doi.org/10.1007/s11119-022-09926-y
Webster, R., & Oliver, M. A. (2007). Geostatistics for environmental scientists (2nd ed.). Wiley. https://doi.org/10.1002/9780470517277
Acknowledgements
This work was funded by the German Federal Ministry of Education and Research (BMBF), Project: “I4S – Intelligence for Soil: Integrated System for Site-specific Soil Fertility Management”, Project No.: 031B1069A and the Agricultural European Innovation Partnership program (EIP-AGRI), Project: “pH-BB: precision liming in Brandenburg”, Project No.: 204016000014/80168341. We are thankful to AMJC Wadoux for his valuable advices concerning the methodology.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conceptualization & methodology: Jonas Schmidinger, Ingmar Schröter, Eric Bönecke, Robin Gebbers, Vera L. Mulder, Gerard B.M. Heuvelink, Sebastian Vogel; Material preparation & data collection: Ingmar Schröter, Eric Bönecke, Robin Gebbers, Joerg Ruehlmann, Eckart Kramer and Sebastian Vogel; Statistical analysis: Jonas Schmidinger; Writing—original draft preparation: Jonas Schmidinger; Review & editing: All authors; Supervision: Sebastian Vogel, Ingmar Schröter, Vera L. Mulder; Funding acquisition: Robin Gebbers, Joerg Ruehlmann, Eckart Kramer.
Corresponding author
Ethics declarations
Competing interest
The authors did not receive support from any organization for the submitted work. The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schmidinger, J., Schröter, I., Bönecke, E. et al. Effect of training sample size, sampling design and prediction model on soil mapping with proximal sensing data for precision liming. Precision Agric 25, 1529–1555 (2024). https://doi.org/10.1007/s11119-024-10122-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-024-10122-3