Abstract
Hyperspectral imaging allows for rapid, non-destructive and objective assessments of crop health. Narrowband-hyperspectral data was used to select wavelength regions that can be exploited to identify wheat infected with soil-borne mosaic virus. First, leaf samples were scanned in the lab to investigate spectral differences between healthy and diseased leaves, including non-symptomatic and symptomatic areas within a diseased leaf. The potential of 84 commonly used vegetation indices to find infection was explored. A machine-learning approach was used to create a classification model to automatically separate pixels into symptomatic, non-symptomatic and healthy classes. The success rate of the model was 69.7% using the full spectrum. It was very encouraging that by using a subset of only four broad bands, sampled to simulate a data set from a much simpler and less costly multispectral camera, accuracy increased to 71.3%. Next, the classification models were validated on field data. Infection in the field was successfully identified using classifiers trained on the entire spectrum of the hyperspectral data acquired in a lab setting, with the best accuracy being 64.9%. Using a subset of wavelengths, simulating multispectral data, the accuracy dropped by only 3 percentage points to 61.9%. This research shows the potential of using lab scans to train classifiers to be successfully applied in the field, even when simultaneously reducing the hyperspectral data to multispectral data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soilborne wheat mosaic virus (SBWMV) poses a threat to the winter wheat industry. In the United States, it was first discovered in the central plains in 1919 (McKinney, 1925). Since then, it has spread, including to northeastern Oregon and southeastern Washington where it threatens the 2-billion-dollar production of dryland winter wheat. Significant reductions of grain yield (40%), biomass (37%) and heads per area (34%) were documented in 2017 and 2018 in association with SBWMV infection in commercial winter wheat fields (Kroese et al., 2020).
The causal agent of SBWMV is a Furovirus which is transmitted by the plasmodiophorid protozoan, Polymyxa graminis (Adams et al., 1988; Estes & Brakke, 1966). Motile zoospores of P. graminis harboring the virus move through the soil solution and are absorbed by root hairs of the plant whereafter they can penetrate cells of the wheat plant, resulting in infection by SBWMV (Slykhuis & Barr, 1978). First symptoms appear in the youngest leaf tissue and spread as the plant grows. Leaf symptoms appear as mottling and are often mistaken for nutrient deficiencies. As the disease progresses, mosaic patches of yellow and green appear on the leaf in a longitudinal pattern along the transverse veins (Kroese et al., 2018). Infection is visible in chlorotic patches in wheat fields, mostly in low-lying areas where standing water supports the swimming zoospore stage of P. graminis. Several species of the wheat genus (Triticum) are impacted, including the commonly grown T. aestivum (Kroese et al., 2018).
Detection of infection currently relies on visual assessment of chlorotic patches in the field. Destructive samples of suspected infection can be molecularly confirmed with PCR-based methods (Kroese et al., 2018). To visualize the distribution of the virus in the field, susceptible varieties of wheat, so called indicator plants, can be planted throughout the field. These indicator plants develop visible symptoms faster than the main variety, intended for harvest and thus presence and spread of infection can easily be monitored.
Current efforts to manage yield loss associated with SBWMV focus on breeding for genetic resistance. However, with early detection of infection, other disease management strategies can be used to minimize yield loss and spread of infection. Such strategies include field isolation, equipment sanitation, overseeding a resistant winter wheat variety, inter-seeding a spring wheat variety and/or terminating the field and planting a profitable non-host crop (e.g. industrial hemp and legume species).
Early and rapid detection of diseases in commercial scale agricultural fields is possible with remote sensing (Zhang et al., 2019), for example by sensors mounted on unmanned aircraft vehicles (UAVs) (Tsouros et al., 2019). Remote sensing is a non-destructive method that uses cameras to measure reflected or emitted light in specific regions of the electromagnetic spectrum, usually reflected light in the visible (VIS), 400–700 nm, and near infra-red (NIR), 700–2500 nm, spectral regions.
Within remote sensing, hyperspectral imaging (HSI) has been widely deployed in plant pathology (Adão et al., 2017; Mahlein, 2016; Mishra et al., 2017; Sahoo et al., 2015). Examples of successful disease detection for wheat include the detection of stem rust (Devadas et al., 2009) and yellow rust (Guo et al., 2021) and the early detection of Fusarium infection (Bauriegel et al., 2011) and powdery mildew (Khan et al., 2021). During HSI, reflectance of 100 + narrow bands is acquired, which results in a continuous spectrum. Typical multispectral cameras acquire only about 5 to 10 broad bands. Note, the distinction between multi- and hyperspectral cameras for sensors with between 11 and 99 bands is arbitrary and more often depends on the width of the measured bands. This increased information from HSI has been more successful at, for example, finding infection in plants. However, HSI comes with a price, both in equipment costs (often > 50,000 USD) and in complexity of subsequent data analysis due to size (typical data acquisition rates are 0.1 Tb/hr from HSI). HSI can therefore be viewed as a specialized research tool and is not geared towards routine applications.
In contrast, multispectral imaging is less expensive and less complex (Moghimi et al., 2018). Programmable multispectral cameras, with different band pass filters, have been applied in wheat studies to track biomass and nitrogen content (Geipel et al., 2016). Wavelength selection for these multispectral cameras often is derived from hyperspectral studies, where scientists strive to find the most informative wavelengths within a hyperspectral data set. For example, Bauriegel et al., (2011) developed a head blight index from a hyperspectral dataset for early detection of Fusarium-infected wheat. For detection of yellow rust (Puccinia striiformis), Bravo et al., (2003) used four bands within the entire hyperspectral dataset. To the authors’ knowledge, no such study has been done for SBWMV.
This research focused on identifying infected areas in wheat fields by SBWMV. The goal was to aid the precision agriculture industry in developing affordable off-the-shelf technology and workflow solutions for the end user by identifying which wavelengths were most informative for SBWMV detection. The foundation for this work was laid using hyperspectral scans of wheat leaves in a laboratory environment to assess discriminatory potential of symptomatic and non-symptomatic wheat leaves. Subsequently, trained models resulting from lab analysis were validated with in-situ scans of field transects.
Methods
Site information
All leaf samples and in-situ measurements were acquired from commercial winter wheat fields in Walla Walla, WA, USA. This location is classified as Köppen’s hot-summer Mediterranean climate zone and experiences average lows of 6 °C, average highs of 18 °C and average rainfall of 530 mm. Soils are predominantly Snow/Athena silt loam and are well drained (Hagerty et al., 2019).
Hyperspectral camera
A Nano VNIR Hyperspec (400–1000 nm) push-broom camera (Headwall Photonics, MA, USA) was used for hyperspectral data acquisition. The camera was fitted with an 8-mm lens with a 30.4° field of view (FOV). It captured data in 271 bands with a 2.2 nm pixel dispersion and 12-bit radiometric resolution with a full width half maximum (FWHM) of 6 nm.
Hyperspectral data acquisition – in-situ scans
In-situ scans were performed to find infection in two SBWMV disease nurseries as well as in two commercial winter wheat fields (Table 1). The disease nurseries had various wheat genotypes for research purposes with different levels of resistance to SBWMV. Nursery one (N1) was labeled healthy and nursery two (N2) was labeled diseased, based on visual assessments and molecular confirmation. In N1, two varieties were targeted: ‘SY Ovation’ (SBWMV resistant) and ‘LCS Artdeco’ (SBWMV semi-resistant). For N2, the same two varieties were scanned along with ‘SG8_92B’ (SBWMV susceptible). Two fields were used for in-field scans. Field one (F1) transects were collected adjacent to N1. Field two (F2) was located elsewhere. Locations in the fields were chosen by experts, based on visual assessments of infected patches (Fig. 1) and follow up molecular confirmation.
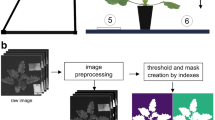
Images were taken using a motion-control system, the Hyper rail (Lopez Alcala et al., 2019). This rail system allows the user to move the camera in a controlled and smooth manner resulting in a coherent image (Fig. 1). Camera settings, such as focus and exposure time, were adjusted according to field conditions. During data acquisition, a 99%-reflectance standard (Labsphere, Inc., North Sutton, NH, USA) was placed in the scene for post-hoc data processing. For every scan, illumination conditions remained stable during the ~ 40-s scan. Images were taken between 2.5 h before and after solar noon to minimize shading within the canopy due to solar angle. Distance from the camera to the wheat plants was 2.5 m, resulting in a spatial resolution of 2 mm and an image swath (width) of 1.3 m.
Hyperspectral data acquisition in situ. (a) Depicts an overview of the Hyper rail with the hyperspectral camera in operation in F2, the chlorotic patch in the field is visible as a lighter green shade (photo credit: Elizabeth Jachens). (b) Is an RGB-composite of the hyperspectral image with a reflectance standard in the scene
Hyperspectral data acquisition – leaf scans
Leaves (n = 105 healthy and 75 diseased, based on visual assessments) were collected on June 6, 2019 from N2 and were shipped overnight in a cooler with icepacks to ensure sample quality. Upon arrival, leaves were immediately scanned under artificial illumination by tungsten halogen lamps. Leaves were placed on a motion-control platform during scanning. Spatial resolution of scans was 0.26 mm per pixel.
Data processing
All image data were radiometrically calibrated using Headwall’s proprietary software, SpectralView (version 5.5.1., Headwall Photonics, MA, USA). The software applies sensor specific response to radiance and a dark reference image. Reflectance was calculated with the empirical line conversion (Smith & Milton, 1999) using the reflectance standard. For lab scans of the leaves, a correction for variation in illumination across the width of the image was applied.
Segmentation
Images were segmented to remove background. For leaf scans, background consisted simply of the platform upon which leaves were scanned. For in-situ scans, background was more complex, where it included soil and other vegetation. A supervised classification algorithm (a support vector machine) was used, trained on visually identified pixels of wheat and background, to rapidly separate wheat-pixels from background-pixels. For both classes, 3000 pixels were used to train all available classifiers in Matlab’s Classification Learner application. The classifier with the highest overall accuracy resulting from a 5-fold cross-validation was identified and consequently used to segment all images. The number of pixels was heuristically determined; more than 3000 pixels did not result in a meaningful increase in visual accuracy but did considerably increase computation time.
Hyperspectral features
Per wheat-pixel, the entire spectrum is extracted. A subset of 84 vegetation indices (VIs) (Online Resource Table S1 from Haagsma et al., (2020)) identified from literature was calculated for each pixel. For these VIs, known correlations exist with plant functioning, which were leveraged to understand impacts of infection. The VIs were organized in sub-categories such as chlorophyll and xanthophyll indices. Furthermore, for leaf-level analysis, not only spectral information was used, but also distribution and texture, i.e., the relationship of a pixel with neighboring pixels. For example, this mosaic virus causes a distinctive spatial discoloring, which can be exploited to find infection. For this analysis, two approaches were used: a measure of distribution of reflectance values per wavelength around the mean, i.e., standard deviation and a measure of spatial autocorrelation, global Moran’s I using Rook’s case adjacency for weight matrix (Moran, 1950).
Training data
Pixels for training data were collected from leaf scans. First, a leaf was randomly selected. From that leaf, pixels were then randomly selected pixels. Within the context of surrounding pixels in the RGB image of the leaf, the pixels were assigned to one of three categories:
-
‘healthy pixel – no symptoms visible on leaf’ (H),
-
‘non-symptomatic pixel from symptomatic (diseased) leaf’ (D-NS),
-
and ‘symptomatic pixel from symptomatic (diseased) leaf’ (D-S)),
hereafter referred to simply as H, D-NS and D-S. For each class, 100 pixels were identified. An exploratory analysis was performed to assess which wavelength had the largest potential to separate three classes by performing a one-way ANOVA test for each wavelength. The p-value resulting from this statistical test is indicative of whether the mean of one of the classes is significantly different from the others. Therefore, this p-value is not directly related to the wavelength’s ability to separate for all three classes. Thus, p-values were compared against the classification accuracies for each of the wavelengths obtained using a support vector machine algorithm. More on classification methods can be found in section ‘Machine learning’.
Feature selection
The most informative VIs were selected based on leaf-pixels using a one-way ANOVA and ranking VIs for p-value (ascending). For wavelength selection, two methods of feature forward selection were used. These methods sequentially select features that contain more information to distinguish between classes.
The first method is a simple correlation and p-value based method (Cp-selection), developed for this study. The Cp-selection is a filter method, i.e., independent from a classification algorithm. First, the p-value from a one-way ANOVA indicates the ability of each feature to separate between three classes. The feature with the smallest p-value is the initial selected feature, identified as the most informative. Next, Pearson’s correlation coefficient between the initial feature and all other features is determined. The next selected feature is the feature with the lowest correlation with the initial feature, providing that its p-value is in the lowest quantile. The correlation matrix is updated with the correlation coefficient of the newest feature with all other features and now contains a column of the initial feature against all others and the second feature against all others. The third, and all features hereafter, are selected based on the smallest total correlation, provided that its p-value is in the lowest quantile. The last steps, starting at updating the correlation matrix, are repeated until a stopping criterion, such as a total number of identified features.
The second feature forward selection method is a built-in algorithm in Matlab, ‘sequentialfs’, here named the FF-selection method. FF-selection is a wrapper method. A wrapper is dependent on a classification algorithm. The selection algorithm determines which candidate feature has the maximum loss in classification error and adds that feature to the selected subset. Here a support vector machine algorithm with a linear kernel was used.
Spectral down-sampling to simulate multispectral data set
Spectral data were averaged over five bands to imitate how selected features will function in a multispectral sensor. With a 2.2 nm distance between the original band centers, this resulted in an 8.8 nm distance between centers of the two outer bands. Considering band width (at half maximum of the signal) of 6 nm, the generated multispectral equivalent data will have a 14.8 nm band width. Similar spectral resolutions (10 ± 2 nm) can be achieved by using a bandpass filter, for example from bk Interferenzoptik Elektronik GmbH (Nabburg, Germany), applied in a multispectral camera (Geipel et al., 2016).
Machine learning
Success of selected features was assessed by using these subsets for classification of leaf-pixels. Classifiers with the highest classification accuracy for (i) all wavelengths and for (ii) a subset of features were selected for in-situ classification.
Two common machine learning algorithms were used to train classifiers based on extracted leaf-pixels: a support vector machine (SVM) and a random forest (RF). Both classifiers are commonly used in studies using hyperspectral data for disease detection in vegetation (Singh et al., 2016). An SVM algorithm searches for the optimal plane in multi-dimensional space to separate data points and is often used in classification problems with a large number of variables and a small number of observations. All the default settings for an SVM with a linear kernel were chosen (kernel is a parameter that determines the shape of the separation plane). RF is an algorithm that ensembles multiple decision trees that are trained to classify data. In this study, 30 learning cycles were used.
For in-situ classification, in addition to SVM and RF, the spectral angle mapper (SAM) was used (Kruse et al., 1993) from Matlab’s Image Processing Toolbox™ Hyperspectral Imaging Library. For SAM, traditionally, endmembers (reference spectra) are extracted from spectral libraries or laboratory scans. Next, these endmembers are used to classify hyperspectral images that have a coarser resolution. Therefore, this algorithm was chosen in addition to SVM and RF, to test the potential of upscaling from lab scans to in-situ. Extracted pixels of leaf scans were used to ensure pure endmembers. It is unlikely to have mixing of target classes within one pixel with a spatial resolution of 0.26 mm. Because endmembers are extracted from leaf scans, SAM was not used to subsequently classify these pixels.
Classification accuracy was estimated by using a 10-fold cross-validation approach (Krstajic et al., 2014). Observations (i.e., the pixels) were divided in 10 subsets. The classifier was trained on nine of these subsets and tested on the out-of-fold subset. This training and testing was repeated 10 times with random subsets created each time and the average accuracy was taken to be the estimated accuracy of the classifier. In a confusion matrix, the classification outcome was presented in terms of the number of occurrences of agreement and disagreement between predicted and true labels of the observations. Moreover, probability of detection (number of accurately identified observations in a class divided by the number of observations that truly fall in that class) was included in the matrix as well as precision (number of accurately identified observations in a class divided by the number of observations predicted to be in that particular class).
In-situ validation
In both nurseries, targeted varieties have known levels of resistance. The level of resistance was leveraged to validate classification outcomes of the nursery scans (Fig. 2). The expectation was that classification outcomes of the SBWMV-resistant variety would show no difference between the healthy (N1) and diseased (N2) nursery. There will be a similar ratio of number of pixels in each class for the healthy and diseased nursery plants. For the semi-resistant variety, it was expected that the ratio of number of pixels in the H class would be larger in the healthy nursery than in the diseased nursery. In conjunction, the number of pixels belonging to D-NS and D-S classes was expected to increase between the healthy and the diseased nursery. For the most susceptible variety, it was expected that the number of pixels in D-NS and D-S classes would be higher than that of resistant and semi-resistant varieties when just considering plants from the diseased nursery.
Expected result of accurate classification of hyperspectral images from the nurseries. ‘Healthy (N1)’ and ‘Diseased (N2)’ labels on the x-axis indicate from which nursery the classification results originate. The y-axis gives the proportion of pixels that fell within the assigned class: H, D-NS and D-S. It was expected that SY Ovation N1, SY Ovation N2 and LCS Artdeco N1 would have a high level of H pixels and low level of D-S pixels, relative to other variety-nursery groupings because N1 was healthy and SY Ovation a resistant variety. Likewise, LCS Artdeco N2 was expected to have a medium level of both H and D-S pixels because LCS Artdeco is semi-resistant and SG8 N2 was expected to have a low level of H and a high level of D-S because of high susceptibility to SBWMV
For validation of the classification of in-field scans, experts’ assessments were used. RGB images, derived from the hyperspectral data, were used to identify areas of infection by two independent expert phytopathologists. Classification outcomes were first spatially smoothed for interpretability using a 15 × 15-pixel moving average. Next, the outcome was visually compared with experts’ assessments in terms of similarity of detected areas of infection. In addition, a pixel based expert assessment was used to quantify the classification accuracy in the field scans. Pixels (n = 140, 20 for each field scan) were randomly chosen for validation and were labeled healthy, symptomatic, or unknown. The asymptomatic class was left out of the validation because this is impossible to assess by visual assessment. The agreement between expert’s assessment and spectral classification was determined for healthy and symptomatic pixels only, the asymptomatic pixels were left out of this analysis. The success of the classifier was reported with accuracy as well as the kappa coefficient. Kappa measures the chance-corrected probability that the classifier agrees with the correct answer. Kappa is normalized to range from 0 (random agreement between true and predicted) to 1 (perfect agreement) (Cohen, 1960). A negative kappa means that the agreement is worse than random.
All data processing and analyses were performed in Matlab R2020b (MathWorks, Natick, MA, USA) unless otherwise stated.
Results and discussion
In the results, the analysis of the lab experiment is presented and discussed. Next, the application of trained classifiers to in-situ data is presented.
Leaf scans
The RGB-composites of healthy and diseased leaves showed differences in symptom expression in the visible spectrum (Fig. 3). The healthy leaf was darker green, which expressed as a lower reflectance in green in the spectral signature (Fig. 4). Strikingly for mosaic virus, the diseased leaf showed streaky patches of infection which is common for mosaic virus (Fig. 3b).
Example of RGB-composite of leaf scans with red markers for pixels used for training data and endmember extraction. Leaf a was healthy, hence all pixels extracted from this leaf were labeled ‘H’ (a1-a3). Leaf b showed symptoms, therefore all extracted pixels were labeled diseased ‘D’. A distinction was made based on close-up images of these pixels between non-symptomatic ‘NS’ (b1 and b3) and symptomatic ‘S’ (b2) pixel
Spectral signatures of the three classes (H, D-NS and D-S) showed differences in both the visible and NIR region of the spectrum (Fig. 4). H and D-NS spectra showed similar reflectance in the visible part of the spectrum. They both had a short peak at the green wavelength (~ 530 nm), indicative of healthy vegetation. However, they diverged in the NIR region. The opposite was true for both classes from diseased leaves, D-NS and D-S. There was a clear difference in the visible region, but a similarity in the NIR region. Reflectance in the NIR region is a function partly of mesophyll sponge structure and has been correlated with biomass depth (thickness of the leaf) (Neuwirthová et al., 2017).
Mean (n = 100) spectral reflectance per class used as training data and endmembers. D-S pixels had a higher reflectivity at the green bump. H and D-NS spectra were similar in the visible part of the spectrum but diverged in the NIR. In contrast, D-NS and D-S pixels had a different reflectance in the visible light, but not in the NIR part of the spectrum. The p-value is of a one-way ANOVA per wavelength. The p-value peaks around the green bump and the red-edge. Spectra at far ends oscillate due to low signal to noise ratio of the sensor
The p-value per wavelength among three classes was most significant for wavelengths around the green peak and at the steep increase from red to NIR, i.e., the red-edge (Fig. 4). Reflectance at 550 nm is sensitive to total chlorophyll content and reflectance at 703 nm is sensitive to vegetation stress (Thenkabail et al., 2004). The log-transformed p-value of the ANOVA was highly correlated (R = -0.87) with the classification accuracy per wavelength and thus p-value was indicative of the wavelength’s ability to separate between the three classes.
The majority of VIs had a significant p-value for dividing pixels into the three classes (Online Resource Table S2). The chlorophyll index red-edge (CI) (Zarco-Tejada et al., 2001) was the VI with the smallest p-value (1.10E-47) (Fig. 5). CI is a simple ratio between reflectance at 750 and 710 nm. The top six identified VIs were all chlorophyll indices (Table S2).
Different VIs have been identified for wheat/pathogen-interaction in previous studies. For example, Devadas et al., (2009) found that the anthocyanin reflectance index (ARI) was best able to discriminate between healthy and rust-infected leaves, while ARI was one of the worst performing indices (63th/84) in this study. However, to track biomass accumulation in winter wheat, Geipel et al., (2016) similarly used chlorophyll-sensitive indices (i.e., normalized difference vegetation index (NDVI) and red-edge inflection point (REIP)).
Classification with full spectrum – leaf level
Using all wavelengths, the classification accuracy was 69.7% for RF and 68.0% for a linear SVM. D-NS pixels had the lowest precision and probability of detection (Table 2) and therefore were the most difficult to identify. The confusion matrix for SVM had similar values (not shown here). The difficulty of D-NS classification might not just be a function of inseparability between spectra, but could also be introduced by errors in manual labeling of pixels. Such errors could be minimized by using assessments of multiple experts. Moreover, an algorithm to identify mislabeled training data can be applied (Brodley & Friedl, 1999).
Classifying with selected features – leaf level
In most cases, a higher classification accuracy was achieved with a larger spectral dataset (Table 3). However, when using only four broad bands, classification accuracy was 71.3% for random forest, compared to 69.7% when using the entire narrow band spectrum. This is a surprising 1.6% point increase in accuracy going from 271 to four bands.
This increase in accuracy is likely caused by spectral averaging of the selected subset. The original accuracy of the classifier trained on four narrow bands was 62.7%. During spectral averaging, noise in the signal is reduced and, in this case, has led to an 8.6 percentage points increase. However, there was no increase in accuracy for the classifier trained on the entire spectrum using broad band spectra. A simple spectral averaging was used to imitate a multispectral-equivalent data set. Each camera has a different sensitivity (bit-depth) and bandwidth-shape (e.g., a sensor measures reflected light in gaussian distribution around a center wavelength). The width and shape of this distribution will directly influence the sensed incoming light. Therefore, in future work, a multispectral camera with these wavelengths needs to be tested on diseased wheat plants. Moreover, other feature selection methods exist, that could further improve accuracy (e.g., in Moghimi et al., 2018).
Texture information – leaf level
Besides looking at absolute reflectance numbers and VIs, texture (distributional and spatial) information was also discriminatory. The p-value of the standard deviation per wavelength had a similar trend as the p-value of the mean reflectance (Figs. 4 and 6). Wavelengths with discriminatory power for absolute reflectance also had discriminatory power in the standard deviation of that wavelength. Moran’s I had significant p-values as well (p < 0.05). However, p-values were larger than p-values of standard deviation and it is therefore expected that they contain less discriminating power. For smaller wavelengths (400–500 nm), Moran’s I did have a smaller p-value than the standard deviation. These texture features will not be transferable to in-situ imagery due to differences in spatial resolution, but they do show promise for analyses at the leaf level.
Plots of the p-value of 2-sample t-tests for healthy and diseased leaves for standard deviation and Moran’s autocorrelation index per wavelength. The p-value of the standard deviation showed a similar pattern to the p-value of the ANOVA tests using the mean reflectance (Fig. 4). For much of the visible part of the spectrum (400–700 nm), both features are significant (p < 0.05)
In-situ results
SBWMV disease nursery
SVM obtained the best classification accuracy (using all wavelengths as well as the selected four bands from FF-selection) (see Fig. 7 caption for more interpretation of the data and Fig. 3 for expected results). SAM with all wavelengths performed equally well. In contrast, RF performed poorly when using either all wavelengths or just the selected bands. SAM with the subset also performed poorly.
Pixel abundance of classification per variety in the healthy (N1) and diseased (N2) nursery. Pixel classes included H (healthy), D-NS (diseased non-symptomatic) and D-S (diseased symptomatic). Panels a, c and e show results using all wavelengths (n = 271). Panels b, d and f show results using four bands selected by FF-selection (multispectral). Panels a and b are results from random forest (RF), panels c and d from the support vector machine (SVM) and panels e and f from the spectral angle mapper (SAM). Panels a, b and f showed unexpected results compared to Fig. 3: they have a large group of D-NS, even in the healthy nursery. In contrast, results in panels c, d and e were expected: the contribution of H pixels was relatively high for varieties in the healthy nursery, as well as in the infected nursery for Ovation and the contribution of H pixels was lower in the infected Artdeco. Contribution of D-S pixels was the highest in the infected SG8 group
Field
Classification results of field scans showed a variation in accuracy (Fig. 8). Here only one field transect is discussed; all other transect-specific results can be found in the supplemental materials (Online Resource Figures S1-S6). The area of infection that was identified by experts on the left side of Fig. 8.a was not classified as symptomatic by any of the classifiers. This area was trampled during installation of equipment and appeared light green in the image because orientation of the leaves was more horizontal compared to surrounding leaves. Chlorotic patches, indicative of SBWMV infection, look similar to these green patches. After closer observation, it is believed that this patch was not showing signs of infection. All classifiers were able to detect that this trampled area was not impacted by disease, while experts, most likely, misclassified this area.
Classification results for F1-T3 (field 1, transect 3). Panel a is an RGB image of the hyperspectral data with expert assessments of infection indicated in black (expert 1) and red (expert 2). The first column with panels b, d and f contains results using all wavelengths, while column 2 with panels c, e and g only used four bands selected with FF-selection. The first row with panels b and c presents results from the RF algorithm, row 2 with d and e from the SVM algorithm and row 3 with f and g from the SAM algorithm. Panels in the first column show similarities with the experts’ assessments, i.e., areas on the right of panel a are also classified diseased (within symptomatic and non-symptomatic range) in panels b, d and f. Areas on the left side of panel a were not apparent in panels b, d and f. Results in column 2 showed less distinct areas of infection for all classifiers. Black indicates soil
RF identified large areas of non-symptomatic infection even where experts’ assessments showed confined symptomatic impact. Therefore, it is believed that RF was unsuccessful at finding infection. SVM found areas with symptoms when using all wavelengths (hyperspectral) but showed only minor success in the four-band classification (multispectral). SAM showed the most distinct patterns compared to all other results when using hyperspectral data. These patterns showed large similarities with the experts’ assessments and therefore SAM was considered accurate. Moreover, extra patches in the top right-corner were identified, which could point at early detection. However, there seems to be an overlap between these symptomatic patches and bare soil. It is known that SBWMV decreases yield and thus one might expect patches of less-dense vegetation. It could also be possible that there was an error in the segmentation of wheat-pixel and background-pixel, resulting in background-pixels being classified as symptomatic wheat. When using multispectral data in SAM, results were unexpected, i.e., overwhelming areas of non-symptomatic and speckles of symptomatic patches.
In all other transect, the same overall principles were apparent. For classifiers using hyperspectral data, all showed similarities with experts’ assessments though RF structurally overestimated the severity of infection. SAM and SVM had similar results, but they were different in sharpness of contrast (SAM had clustered areas of symptoms where SVM showed more bleeding of non-symptomatic into healthy areas and vice versa). For results using a subset of four bands, SVM under-estimated the infection, while both RF and SAM over-estimated the extent and severity of infection.
For pixel-wise validation, SVM with hyperspectral data had the highest accuracy (Table 4). However, the accuracies and kappa coefficients of the classifiers for hyperspectral data were very similar; there was no superior classifier. For multispectral data, the best accuracy was for SVM and it was only 3 percentage points lower than SVM’s hyperspectral accuracy. Even though the accuracies in multispectral data was the same for SVM and RF, SVM had a bigger kappa coefficient and therefore is the superior classifier.
Thus, based on the classification of field scans, it is recommended to use SAM or SVM with all wavelengths and SVM when only using the subset of 4 bands. The nursery results corroborate this finding.
Conclusion
Lab scans of healthy leaves (leaves without signs of SBWMV infection) and SBWMV-infected leaves were used to extract hyperspectral features of interest. It was found that the CI, within a subset of VIs, had the largest ability to separate for the three target classes (healthy, diseased-non-symptomatic and diseased-symptomatic). CI is a simple band ratio between 750 and 710 nm and is sensitive to the steepness of the red-edge. Other VIs of interest were all sensitive to changes in chlorophyll content. Using CI, a leaf-pixel classification accuracy of 65.3% was achieved, while using all wavelengths resulted in a 69.7% accuracy. A subset of wavelengths was selected with a feature forward selection. These wavelengths were converted to multispectral equivalent broad band reflectance data. Using these data increased the classification accuracy to 71.3%. During spectral averaging, noise in the signal is reduced and, in this case, led to an 8.6percentage points increase in accuracy.
Classifiers trained on lab-scan data were applied in the field. It was shown that upscaling to larger pixels might be possible, but that more research is needed. The most successful classifiers in terms of identifying patches of infection similar to expert assessments were SVM and SAM when using the entire spectrum. Pixel-wise validation estimated the accuracy of healthy and symptomatic classification to be 63.4% (SAM) and 64.9% (SVM). Using the best subset, derived from data analysis of the lab scans, patterns of infection were less pronounced. SVM showed promising results with an estimated accuracy of 61.9%, only percentage points lower than the hyperspectral equivalent. Further research should explore the potential of the selected wavelengths in a multispectral camera in other field settings. In addition, the potential of these wavelengths to distinguish between different stresses in the field should be investigated. In future work, this concept may be adapted to other pathosystems and the methods presented in this paper should be tested in fields beyond plant pathology, where there is a need for rapid and reliable assessment without the possibility of continuous deployment of a hyperspectral camera.
Availability of data and material
The datasets generated during and/or analyzed during the current study are available in the Oregon State University’s Scholars Archive repository, https://doi.org/10.7267/z316q855z.
Code availability
Code will be made available upon request.
References
Adams, M. J., Swaby, A. G., & Jones, P. (1988). Confirmation of the transmission of barley yellow mosaic virus (BaYMV) by the fungus polymyxa graminis. Annals of Applied Biology, 112(1), 133–141. https://doi.org/10.1111/j.1744-7348.1988.tb02048.x.
Adão, T., Hruška, J., Pádua, L., Bessa, J., Peres, E., Morais, R., et al. (2017). Hyperspectral imaging: a review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sensing, 9(11), https://doi.org/10.3390/rs9111110.
Bauriegel, E., Giebel, A., Geyer, M., Schmidt, U., & Herppich, W. B. (2011). Early detection of Fusarium infection in wheat using hyper-spectral imaging. Computers and Electronics in Agriculture, 75(2), 304–312. https://doi.org/10.1016/j.compag.2010.12.006.
Bravo, C., Moshou, D., West, J., McCartney, A., & Ramon, H. (2003). Early Disease Detection in Wheat Fields using Spectral Reflectance. Biosystems Engineering, 84(2), 137–145. https://doi.org/10.1016/S1537-5110(02)00269-6.
Brodley, C. E., & Friedl, M. A. (1999). Identifying mislabeled Training Data. Journal of Artificial Intelligence Research, 11, 131–167. https://doi.org/10.1613/jair.606.
Cohen, J. (1960). A coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement, 20(1), 37–46. https://doi.org/10.1177/001316446002000104.
Devadas, R., Lamb, D. W., Simpfendorfer, S., & Backhouse, D. (2009). Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precision Agriculture, 10(6), 459–470. https://doi.org/10.1007/s11119-008-9100-2.
Estes, A., & Brakke, M. (1966). Correlation of Polymyxa graminis with transmission of soil-borne wheat mosaic virus. Virology, 28(4), 772–774. https://www.sciencedirect.com/science/article/pii/0042682266902662.
Geipel, J., Link, J., Wirwahn, J. A., & Claupein, W. (2016). A programmable aerial multispectral camera system for in-season crop biomass and nitrogen content estimation. Agriculture, 6(1), 1–19. https://doi.org/10.3390/agriculture6010004.
Guo, A., Huang, W., Dong, Y., Ye, H., Ma, H., Liu, B., et al. (2021). Wheat yellow rust detection using UAV-Based Hyperspectral Technology. Remote Sensing, 1313(1), 123. https://doi.org/10.3390/RS13010123.
Haagsma, M., Page, G. F. M., Johnson, J. S., Still, C., Waring, K. M., Sniezko, R. A., et al. (2020). Using hyperspectral imagery to detect an invasive fungal pathogen and symptom severity in pinus strobiformis seedlings of different genotypes. Remote Sensing, 12(24), 1–19. https://doi.org/10.3390/rs12244041.
Hagerty, C. H., Fickas, K. C., & Wysocki, D. (2019). Agronomic Zones of the Dryland Pacific Northwest. PNW 354, March. A Pacific Northwest Extension Publication. University of Idaho Cooperative Extension System, Oregon State University Extension Service and the Washington State university Cooperative Extension System. Retrieved 26 May 2021 from https://catalog.extension.oregonstate.edu/pnw354
Khan, I. H., Liu, H., Li, W., Cao, A., Wang, X., Liu, H., et al. (2021). Early detection of Powdery Mildew Disease and Accurate quantification of its severity using hyperspectral images in wheat. Remote Sensing, 13(18), 3612. https://doi.org/10.3390/RS13183612.
Kroese, D. R., Bag, S., Frost, K. E., Murray, T. D., & Hagerty, C. H. (2018). Diagnostic Guide: Wheat Soil-Borne Mosaic. Plant Health Progress, 19(2), 163–167. https://doi.org/10.1094/PHP-10-17-0064-DG.
Kroese, D. R., Schonneker, L., Bag, S., Frost, K., Cating, R., & Hagerty, C. H. (2020). Wheat soil-borne mosaic: yield loss and distribution in the US Pacific Northwest. Crop Protection, 132, 105102. https://doi.org/10.1016/j.cropro.2020.105102.
Krstajic, D., Buturovic, L. J., Leahy, D. E., & Thomas, S. (2014). Cross-validation pitfalls when selecting and assessing regression and classification models. Journal of Cheminformatics, 6 10. https://doi.org/10.1186/1758-2946-6-10
Kruse, F. A., Lefkoff, A. B., Boardman, J. W., Heidebrecht, K. B., Shapiro, A. T., Barloon, P. J., et al. (1993). The spectral image Processing System (SIPS) - interactive visualization and analysis of imaging Spectrometer Data. Remote Sensing of Environment, 44(2–3), 145–163. https://doi.org/10.1016/0034-4257(93)90013-N.
Lopez Alcala, J. M., Haagsma, M., Udell, C. J., & Selker, J. S. (2019). HyperRail: modular, 3D printed, 1-100 meter, programmable, and low-cost Linear Motion Control System for Imaging and Sensor Suites. HardwareX, e00081. https://doi.org/10.1016/j.ohx.2019.e00081.
Mahlein, A. K. (2016). Plant Disease Detection by Imaging Sensors - Parallels and Specific demands for Precision Agriculture and Plant phenotyping. Plant Disease, 100(2), 241–251. https://doi.org/10.1094/PDIS-03-15-0340-FE.
McKinney, H. (1925). Mosaic disease of winter wheat and winter rye. Retrieved 20 January 2021 from http://agris.fao.org/agris-search/search.do?recordID=US201300429200
Mishra, P., Asaari, M. S. M., Herrero-Langreo, A., Lohumi, S., Diezma, B., & Scheunders, P. (2017). Close range hyperspectral imaging of plants: a review. Biosystems Engineering, 164, 49–67. https://doi.org/10.1016/j.biosystemseng.2017.09.009.
Moghimi, A., Yang, C., & Marchetto, P. M. (2018). Ensemble feature selection for plant phenotyping: a journey from Hyperspectral to Multispectral Imaging. Ieee Access : Practical Innovations, Open Solutions, 6, 56870–56884. https://doi.org/10.1109/ACCESS.2018.2872801.
Moran, P. A. P. (1950). Notes on continuous stochastic phenomena. Biometrika, 37(1/2), 17. https://doi.org/10.2307/2332142.
Neuwirthová, E., Lhotáková, Z., & Albrechtová, J. (2017). The effect of leaf stacking on leaf reflectance and vegetation indices measured by contact probe during the season. Sensors (Basel, Switzerland), 17(6), 1202. https://doi.org/10.3390/s17061202.
Sahoo, R. N., Ray, S. S., & Manjunath, K. R. (2015). Hyperspectral remote sensing of agriculture. Current Science, 108(5), 848–859. https://doi.org/10.1201/b11222.
Singh, A., Ganapathysubramanian, B., Singh, A. K., & Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends in Plant Science, 21(2), 110–124. https://doi.org/10.1016/j.tplants.2015.10.015.
Slykhuis, J. T., & Barr, D. J. S. (1978). Confirmation of Polymyxa graminis as a Vector of Wheat Spindle Streak Mosaic Virus. Phytopathology, 68(4), 639–643. https://doi.org/10.1094/phyto-68-639.
Smith, G., & Milton, E. (1999). The use of the empirical line method to calibrate remotely sensed data to reflectance. International Journal of Remote Sensing, 20(13), 2653–2662. https://doi.org/10.1080/014311699211994.
Thenkabail, P. S., Enclona, E. A., Ashton, M. S., & Van Der Meer, B. (2004). Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sensing of Environment, 91(3–4), 354–376. https://doi.org/10.1016/j.rse.2004.03.013.
Tsouros, D. C., Bibi, S., & Sarigiannidis, P. G. (2019). A review on UAV-Based applications for Precision Agriculture. Information, 10(11), 349. https://doi.org/10.3390/info10110349.
Zarco-Tejada, P. J., Miller, J. R., Noland, T. L., Mohammed, G. H., & Sampson, P. H. (2001). Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Transactions on Geoscience and Remote Sensing, 39(7), 1491–1507. https://doi.org/10.1109/36.934080.
Zhang, J., Huang, Y., Pu, R., Gonzalez-Moreno, P., Yuan, L., Wu, K., et al. (2019). Monitoring plant diseases and pests through remote sensing technology: a review. Computers and Electronics in Agriculture, 165, 104943. https://doi.org/10.1016/j.compag.2019.104943.
Acknowledgements
We thank the faculty, staff and student researchers at the Columbia Basin Agriculture Research Center for supporting this research project. We thank the Oregon Wheat Commission and the growers who kindly allowed us to conduct research on their farms. In addition, we thank Elizabeth Jachens for her diligent notetaking during fieldwork and the OPEnS lab for their support on preparing the Hyper rail for field work. And many thanks to Gordon Godshalk for proofreading the article.
Funding
Funding for this project was provided by the Agriculture Research Foundation competitive grant program and by the National Science Foundation under grant nr. NSF-EAR 1440506 and NSF-CCF 1521687 (Collaborative Research: Facility Support: Center for Transformative Environmental Monitoring Programs (CTEMPs); and Collaborative Research; CompSustNet: Expanding the Horizons of Computational Sustainability, respectively).
Author information
Authors and Affiliations
Contributions
MH: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing, Visualization. CHH: Conceptualization, Methodology, Investigation, Resources, Writing – Original Draft, Writing – Review & Editing, Supervision, Project administration, Funding acquisition. DRK: Investigation, Resources, Writing – Review & Editing. JSS: Conceptualization, Resources, Writing – Review & Editing, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Conflicts of interest/competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haagsma, M., Hagerty, C.H., Kroese, D.R. et al. Detection of soil-borne wheat mosaic virus using hyperspectral imaging: from lab to field scans and from hyperspectral to multispectral data. Precision Agric 24, 1030–1048 (2023). https://doi.org/10.1007/s11119-022-09986-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-022-09986-0