Abstract
Aquaporins (APQs) belong to the major intrinsic protein superfamily and play a key role in the transport of water and other solutes across cell membranes. Coffea canephora is an evergreen shrub used for making instant coffees. Genome analysis of C. canephora identified 33 putative aquaporin genes assigned to five subfamilies including seven plasma membrane intrinsic proteins (PIP), 9 tonoplast intrinsic proteins (TIP), 11 NOD26-like intrinsic proteins (NIP), 3 small basic intrinsic proteins (SIP), and 3 X intrinsic proteins (XIP). Generally, the AQPs gene structure was conserved within each subfamily, with exon numbers ranging from one to five. The prediction of the aromatic/arginine selectivity filter (ar/R) and Froger’s positions indicated a noticeable difference in substrate specificity between subfamilies. Synteny analysis revealed high conservation of aquaporin genes in coffee. In silico expression analysis of the CcAQPs genes indicated that they were differentially expressed in various tissues. Members of CcPIPs and CcTIPs subfamilies were validated by real-time quantitative analysis in leaves of two genotypes of C. canephora with contrasting responses to water deficit (clone 14: drought-tolerant and clone 109A: drought-susceptible). Under severe water deficit, the relative expression of isoforms of both genes decreased in clone 14 compared with that under the irrigated condition, while clone 109A showed comparatively higher mRNA levels, with the exception of CcPIP1;2 in the stress condition. This study was the first to characterize and validate aquaporin genes in C. canephora in response to water deficit, and the findings may provide insights for biotechnological approaches to increase tolerance to drought.







Similar content being viewed by others
References
Afzal Z, Howton TC, Sun Y, Shahid Mukhtar M (2016) The roles of aquaporins in plant stress responses. J Dev Biol 4:1–9. https://doi.org/10.3390/jdb4010009
Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484. https://doi.org/10.1007/s11103-005-0352-1
Alexandersson E, Danielson JAH, Rade J, Moparthi VK, Fontes M, Kjellbom P, Johanson U (2010) Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J 61:650–660. https://doi.org/10.1111/j.1365-313X.2009.04087.x
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Anderberg HI, Kjellbom P, Johanson U (2012) Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front Plant Sci 3:33. https://doi.org/10.3389/fpls.2012.00033
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44:147–153. https://doi.org/10.1093/nar/gkw419
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:369–373. https://doi.org/10.1093/nar/gkl198
Bari A, Farooq M, Hussain A, Qamar MT, Abbas MW, Mustafa G, Karim A, Ahmed I, Hussain T (2018) Genome-wide bioinformatics analysis of aquaporin gene family in maize (Zea mays L.). J Phylogenet Evol Biol 6:197. https://doi.org/10.4172/2329-9002.1000197
Bienert GP, Møller AL, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282:1183–1192. https://doi.org/10.1074/jbc.M603761200
Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F (2011) Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J 66:306–317. https://doi.org/10.1111/j.1365-313X.2011.04496.x
Bienert MD, Bienert GP (2017) Plant Aquaporins and metalloids. In: Chaumont F, Tyerman S (eds) Plant aquaporins signaling and communication in plants. Springer, Cham, pp 297–332
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10. https://doi.org/10.1186/1471-2229-4-10
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. https://doi.org/10.1371/journal.pone.0011335
Da Matta FM, Ramalho JC (2006) Impacts of drought and temperature stress on coffee physiology and production: a review. Braz J Plant Physiol 18:55–81. https://doi.org/10.1590/S167704202006000100006
Danielson JÅH, Johanson U (2008) Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol 8:45. https://doi.org/10.1186/1471-2229-8-45
Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, Aury JM, Bento P, Bernard M, Bocs S, Campa C, Cenci A, Combes MC, Crouzillat D, da Silva C, Daddiego L, de Bellis F, Dussert S, Garsmeur O, Gayraud T, Guignon V, Jahn K, Jamilloux V, Joet T, Labadie K, Lan T, Leclercq J, Lepelley M, Leroy T, Li LT, Librado P, Lopez L, Munoz A, Noel B, Pallavicini A, Perrotta G, Poncet V, Pot D, Priyono, Rigoreau M, Rouard M, Rozas J, Tranchant-Dubreuil C, VanBuren R, Zhang Q, Andrade AC, Argout X, Bertrand B, de Kochko A, Graziosi G, Henry RJ, Jayarama, Ming R, Nagai C, Rounsley S, Sankoff D, Giuliano G, Albert VA, Wincker P, Lashermes P (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184. https://doi.org/10.1126/science.1255274
Deokar AA, Tar’An B (2016) Genome-wide analysis of the aquaporin gene family in chickpea (Cicer arietinum L.). Front Plant Sci 7:1802. https://doi.org/10.3389/fpls.2016.01802
Deshmukh R, Bélanger RR (2016) Molecular evolution of Aquaporins and silicon influx in plants. Funct Ecol 30:1277–1285. https://doi.org/10.1111/1365-2435.12570
Diehn TA, Pommerrenig B, Bernhardt N, Hartmann A, Bienert GP (2015) Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front Plant Sci 6:166. https://doi.org/10.3389/fpls.2015.00166
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183:557–564. https://doi.org/10.1111/j.1469-8137.2009.02923.x
Forrest KL, Bhave M (2007) Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct Integr Genomics 7:263–289. https://doi.org/10.1007/s10142-007-0049-4
Froger A, Thomas D, Delamarche C, Tallur B (1998) Prediction of functional residues in water channels and related proteins. Protein Sci 7:1458–1468. https://doi.org/10.1002/pro.5560070623
Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor–like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18:2021–2034. https://doi.org/10.1105/tpc.106.043000
Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F (2009) Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta 1788:1213–1228. https://doi.org/10.1016/j.bbamem.2009.03.009
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:1178–D1186. https://doi.org/10.1093/nar/gkr944
Gupta AB, Sankararamakrishnan R (2009) Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol 9:134. https://doi.org/10.1186/1471-2229-9-134
Heymann JB, Engel A (2000) Structural clues in the sequences of the aquaporins. J Mol Biol 295:1039–1053. https://doi.org/10.1006/jmbi.1999.3413
Hooijmaijers C, Rhee JY, Kwak KJ, Chung GC, Horie T, Katsuhara M, Kang H (2012) Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. J Plant Res 125:147–153. https://doi.org/10.1007/s10265-011-0413-2
Hove RM, Bhave M (2011) Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol 75:413–430. https://doi.org/10.1007/s11103-011-9737-5
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Huang Z, Zhong XJ, He J, Jin SH, Guo HD, Yu XF, Zhou YJ, Li X, Ma MD, Qi-Bing Chen QB, Long H (2016) Genome-wide identification, characterization, and stress-responsive expression profiling of genes encoding LEA (late embryogenesis abundant) proteins in moso bamboo (Phyllostachys edulis). PLoS One 11:e0165953. https://doi.org/10.1371/journal.pone.0165953
Hub JS, de Groot BL (2008) Mechanism of selectivity in aquaporins and aquaglyceroporins. PNAS 105:1198–1203. https://doi.org/10.1073/pnas.0707662104
Hussain A, Tanveerc R, Mustafa G, Farooq M, Amina I, Mansoora S (2019) Comparative phylogenetic analysis of aquaporins provides insight into the gene family expansion and evolution in plants and their role in drought tolerant and susceptible chickpea cultivars. Genomics 112:263–275. https://doi.org/10.1016/j.ygeno.2019.02.005
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126:1358–1369. https://doi.org/10.1104/pp.126.4.1358
Johanson U, Gustavsson S (2002) A new subfamily of major intrinsic proteins in plants. Mol Biol Evol 19:456–461. https://doi.org/10.1093/oxfordjournals.molbev.a004101
Kadam S, Abril A, Dhanapal AP, Koester RP, Vermerris W, Jose S, Fritschi FB (2017) Characterization and Regulation of Aquaporin Genes of Sorghum [Sorghum bicolor (L.) Moench] in Response to Waterlogging Stress. Frontiers in Plant Science 8:862. https://doi.org/10.3389/fpls.2017.00862
Khan K, Agarwal P, Shanware A, Sane VA (2015) Heterologous expression of two jatropha aquaporins imparts drought and salt tolerance and improves seed viability in transgenic Arabidopsis thaliana. PLoS One 10:e0128866. https://doi.org/10.1371/journal.pone.0128866
Kong W, Yang S, Wang Y, Bendahmane M, Fu X (2017) Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. PeerJ 5:e3747. https://doi.org/10.7717/peerj.3747
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra M (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGAX: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lashermes P, Combes MC, Robert J, Troulot P, D’Hont A, Anthony F, Charrier A (1999) Molecular characterization and origin of the Coffea arabica L. genome. Mol Gen Genet 261:259–266. https://doi.org/10.1007/s004380050
Li T, Choi WG, Wallace IS, Baudry J, Roberts DM (2011) Arabidopsis thaliana NIP7;1: an anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 50(31):6633–6641. https://doi.org/10.1021/bi2004476
Li G, Santoni V, Maurel C (2014) Plant aquaporins: roles in plant physiology. BBA Gen Subj 1840:1574–1582. https://doi.org/10.1007/s004380050
Lima EA, Furlanetto C, Nicole M, Gomes AC, Almeida MR, Jorge-Júnior A, Correa VR, Salgado SM, Ferrão MA, Carneiro RM (2015) The multi-resistant reaction of drought-tolerant coffee ‘Conilon clone 14’to Meloidogyne spp. and late hypersensitive-like response in Coffea canephora. Phytopathology 105:805–814. https://doi.org/10.1094/PHYTO-08-14-0232-R
Liu Q, Wang H, Wu J, Zang Z, Wu J, Feng Y, Zhu Z (2009) Divergence in function and expression of the NOD26-like intrinsic proteins in plants. BMC Genomics 10:313. https://doi.org/10.1186/1471-2164-10-313
Liu LH, Ludewig U, Gassert B, Frommer WB, Von Wirén N (2003) Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol 133:1220–1228. https://doi.org/10.1104/pp.103.027409
Liu Q, Zhu Z (2010) Functional divergence of the NIP III subgroup proteins involved altered selective constraints and positive selection. BMC Plant Biol 10:256. https://doi.org/10.1186/1471-2229-10-256
Loqué D, Ludewig U, Yuan L, von Wirén N (2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol 137:671–680. https://doi.org/10.1104/pp.104.051268
Mao Z, Sun W (2015) Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. J Exp Bot 66:4781–4794. https://doi.org/10.1093/jxb/erv244
Marraccini P, Vinecky F, Alves GSC, Ramos HJO, Elbelt S, Vieira NG, Carneiro FA, Sujii PS, Alekcevetch JC, Silva VA, DaMatta FM, Ferrao MAG, Leroy T, Pot D, Vieira LGE, da Silva FR, Andrade AC (2012) Differentially expressed genes and proteins upon drought acclimation in tolerant and sensitive genotypes of Coffea canephora. J Exp Bot 63:4191–4212. https://doi.org/10.1093/jxb/ers103
Martins CPS, Pedrosa AM, Du D, Gonçalves LP, Yu Q, Gmitter JFG, Costa MG (2015) Genome-wide characterization and expression analysis of major intrinsic proteins during abiotic and biotic stresses in sweet orange (Citrus sinensis L. Osb.). PLoS One 10:e0138786. https://doi.org/10.1371/journal.pone.0138786
Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucp L (2015) Aquaporins in plants. Physiol Rev 95:1321–1358. https://doi.org/10.1152/physrev.00008.2015
Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF (2011) The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot 62:4391–4398. https://doi.org/10.1093/jxb/err158
Miniussi M, , Savi TL, Pallavicini A, Nardini A (2015) Aquaporins in Coffea arabica L.: identification, expression, and impacts on plant water relations and hydraulics. Plant Physiol Biochem95:92–102. https://doi.org/10.1016/j.plaphy.2015.07.024
Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y (2000) Structural determinants of water permeation through aquaporin-1. Nature 407:599–605. https://doi.org/10.1038/35036519
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):2002–2007. https://doi.org/10.1093/nar/29.9.e45
Park W, Scheffler BE, Bauer PJ, Campbell BT (2010) Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol 10:142. https://doi.org/10.1186/1471-2229-10-142
Perez-Martin A, Michelazzo C, Torres-Ruiz JM, Flexas J, Fernández JE, Sebastiani L, Diaz-Espejo A (2014) Regulation of photosynthesis and stomatal and mesophyll conductance under water stress and recovery in olive trees: correlation with gene expression of carbonic anhydrase and aquaporins. J Exp Bot 65:3143–3156. https://doi.org/10.1093/jxb/eru160
Pommerrenig B, Diehn TA, Bienert GP (2015) Metalloido-porins: essentiality of nodulin 26-like intrinsic proteins in metalloid transport. Plant Sci 238:212–227. https://doi.org/10.1016/j.plantsci.2015.06.002
Ramakers C, Ruijtera JM, Depreza RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. https://doi.org/10.1016/S0304-3940(02)01423-4
Reddy PS, Rao TRRB, Sharma KK, Vadez V (2015) Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L). Plant Gene 1:18–28. https://doi.org/10.1016/j.plgene.2014.12.002
Reuscher S, Akiyama M, Mori C, Aoki K, Shibata D, Shiratake K (2013) Genome-wide identification and expression analysis of aquaporins in tomato. Plos One 8(11):e79052. https://doi.org/10.1371/journal.pone.0079052
Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46:1568–1577. https://doi.org/10.1093/pcp/pci172
Santos AB, Mazzafera P (2013) Aquaporins and the control of the water status in coffee plants. Theor Exp Plant Physiol 25:79–93. https://doi.org/10.1590/S2197-00252013000200001
Santos TB, Lima RBL, Nagashima GT, Petkowicz CLO, Carpentieri-Pípolo V, Pereira LPP, Domingues DS, Vieira LGE (2015) Galactinol synthase transcriptional profile in two genotypes of Coffea canephora with contrasting tolerance to drought. Genet Mol Biol 38:182–190. https://doi.org/10.1590/S1415-475738220140171
Shekoofa A, Sinclair T (2018) Aquaporin activity to improve crop drought tolerance. Cells 7:123. https://doi.org/10.3390/cells7090123
Shivaraj SM, Deshmukh RK, Rai R, Bélanger R, Agrawal PK, Dash PK (2017) Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci Rep 7:46137. https://doi.org/10.1038/srep4613
Sun H, Wang S, Lou Y, Zhu C, Zhao H, Li Y, Li X, Gao Z (2018) Whole-genome and expression analyses of bamboo aquaporin genes reveal their functions involved in maintaining diurnal water balance in bamboo shoots. Cells 7:195. https://doi.org/10.3390/cells7110195
Takano J, Motoko W, Ludewig U, Schaaf G, von Wirén N, Fujiwara T (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18:498–1509. https://doi.org/10.1105/tpc.106.041640
Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T (2008) NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20:2860–2875. https://doi.org/10.1105/tpc.108.058628
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P (2006) Structural mechanism of plant aquaporin gating. Nature 439:688–694. https://doi.org/10.1038/nature04316
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wei Q, Ma Q, Ma Z, Zhou G, Feng F, Le S, Lei C, Gu Q (2019) Genome-wide identification and characterization of sweet orange (Citrus sinensis) aquaporin genes and their expression in two citrus cultivars differing in drought tolerance. Tree Genet Genomes 15:17. https://doi.org/10.1007/s11295-019-1321-1
Yakata K, Hiroaki Y, Ishibashi K, Sohara E, Sasaki S, Mitsuoka K, Fujiyoshi Y (2007) Aquaporin-11 containing a divergent NPA motif has normal water channel activity. Biochim Biophys Acta 1768:688–693. https://doi.org/10.1016/j.bbamem.2006.11.005
Yu Q, Guyot R, de Kochko A, Byers A, Navajas-Pérez R, Langston BJ, Dubreuil-Tranchant C, Paterson AH, Poncet V, Nagai C, Ming R (2011) Micro-collinearity and genome evolution in the vicinity of an ethylene receptor gene of cultivated diploid and allotetraploid coffee species (Coffea). Plant J 67:305–317. https://doi.org/10.1111/j.1365-313X.2011.04590.x
Zhang Z, Li J, Yu J (2006) Computing Ka and Ks with a consideration of unequal transitional substitutions. BMC Evol Biol 6:44. https://doi.org/10.1186/1471-2148-6-44
Zhang B, Xie L, Sun T, Ding B, Li Y, Zhang Y (2019) Chrysanthemum morifolium aquaporin genes CmPIP1 and CmPIP2 are involved in tolerance to salt stress. Sci Hortic 256:108627. https://doi.org/10.1016/j.scienta.2019.108627
Zhu YX, Yang L, Liu N, Yang J, Zhou XK, Xia YC, He Y, He YQ, Gong HJ, Ma DF, Yin JL (2019) Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biology 19:345. https://doi.org/10.1186/s12870-019-1953-1
Zardoya R, Villalba S (2001) A phylogenetic framework for the aquaporin family in eukaryotes. J Mol Evol 52:391–404. https://doi.org/10.1007/s002390010169
Zargar SM, Nagar P, Deshmukh R, Nazir M, Wani AA, Masoodi KZ, Agrawal GK, Rakwal R (2017) Aquaporins as potential drought tolerance inducing proteins: towards instigating stress tolerance. J Proteome 169:233–238. https://doi.org/10.1016/j.jprot.2017.04.010
Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH (2009) Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet 5:e1000581. https://doi.org/10.1371/journal.pgen.1000581
Zou Z, Gong J, Huang Q, Mo Y, Yang L, Xie G (2015) Gene structures, evolution, classification and expression profiles of the aquaporin gene family in castor bean (Ricinus communis L.). Plos One 10:e0141022. https://doi.org/10.1371/journal.pone.0141022
Zupin M, Sedlar A, Kidrič M, Meglič V (2017) Drought-induced expression of aquaporin genes in leaves of two common bean cultivars differing in tolerance to drought stress. J Plant Res 130:735–745. https://doi.org/10.1007/s10265-017-0920-x
Funding
The author Diliane Harumi Yaguinuma received a grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Grant Number 001).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
•Aquaporin gene family conservation maintained in coffee plants. PIPs and TIPs isoforms expressed differentially in the contrasting genotypes of Coffea canephora in response to water deficit.
Electronic Supplementary Material
Fig. S1
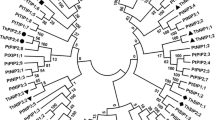
Phylogenetic tree of AQPs of C. canephora and A. thaliana using the standard ClustalW alignment parameter. The phylogenetic dendrogram generated by MEGAX using the ML method. Bootstrap analysis was performed with 1000 replicates; only bootstrap values higher than 50% are indicated above the branches. (PNG 157 kb)
Fig. S2
Partial alignment of putative XIP sequences of three Coffea species (C. arabica (Ca), C. eugenioides (Ce) and C. canephora (Cc) using the ClustalW tool through the MEGAX platform. Highlighted in red is the NPARC motif. (PNG 49 kb)
Fig. S3
Multiple alignment of deduced amino acid sequences of CcAQP using the ClustalW tool available in the MEGAX. NPA motifs (bold); ar / R selectivity filters (H2, H5, LE1 and LE2) are highlighted in red; Froger’s 5 positions (P1 - P5) highlighted in green. Underlined transmembrane helix positions. (DOCX 125 kb)
Fig. S4
Prediction of transmembrane domains in the deduced amino acid sequences of CcAQP through the TMHMM Server v. 2.0. (DOCX 716 kb)
Fig. S5
Phylogenetic tree of C. arabica and A. thailana using the CLUSTALW standard alignment parameter. The phylogenetic dendrogram generated by MEGA X using the Maximum Likelihood method with 1000 bootstrap replicates (PNG 117 kb)
Fig. S6
Phylogenetic tree of C. eugenioides and A. thailana using the standard CLUSTALW alignment parameters. The phylogenetic dendrogram generated by MEGA X using the Maximum Likelihood method with 1000 bootstrap replicates. (PNG 106 kb)
Table S1
Date of duplication of the pairs of paralogous genes of the C. canephora aquaporin gene family. Ka represents the non-synonymous substitution number per non-synonymous site, Ks is the number of the synonymous substitution site; Ka/Ks represents the ratio of non-synonymous (Ka) to synonymous (Ks) substitutions. (DOCX 16 kb)
Table S2
Aquaporin genes synteny among three Coffea species (C. arabica (Ca), C. eugenioides (Ce) and C. canephora (Cc). (DOCX 19 kb)
Table S3
Aquaporin genes synteny between Coffea arabica (Ca) and Coffea canephora (Cc). (DOCX 19 kb)
Table S4
Aquaporin genes synteny between C. eugenioides (Ce) and C. canephora (Cc). (DOCX 17 kb)
Table S5
Primers, amplicon size and amplification efficiency for RT-qPCR analysis of CcAQPs. (DOCX 15 kb)
Table S6
List of accession numbers of from all aquaporin genes used for phylogenetic analysis (DOCX 40 kb)
Rights and permissions
About this article
Cite this article
Yaguinuma, D.H., dos Santos, T.B., de Souza, S.G.H. et al. Genome-Wide Identification, Evolution, and Expression Profile of Aquaporin Genes in Coffea canephora in Response to Water Deficit. Plant Mol Biol Rep 39, 146–162 (2021). https://doi.org/10.1007/s11105-020-01235-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01235-w