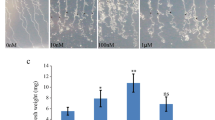
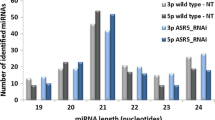
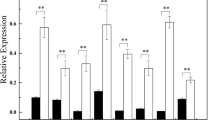
Abstract
MicroRNAs (miRNAs) are a class of small endogenous RNAs conserved in eukaryotic organisms including plants. They suppress gene expression posttranscriptionally in many different biological processes. Previously, we reported salinity-induced changes in gene expression in transgenic Arabidopsis thaliana plants that constitutively expressed a pea abscisic acid-responsive (ABR17) gene. In the current study, we used microarrays to investigate the role of miRNA-mediated posttranscriptional gene regulation in these same transgenic plants in the presence and absence of salinity stress. We identified nine miRNAs that were significantly modulated due to ABR17 gene expression and seven miRNAs that were modulated in response to salt stress. The target genes regulated by these miRNAs were identified using sRNA target Base (starBase) degradome analysis and through 5′-RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE). Our findings revealed miRNA–mRNA interactions comprising regulatory networks of auxin response factor (ARF), argonaute 1 (AGO1), dicer-like proteins 1 (DCL1), SQUAMOSA promoter binding (SPB), NAC, APETALA 2 (AP2), nuclear factor Y (NFY), RNA-binding proteins, A. thaliana vacuolar phyrophosphate 1 (AVP1), and pentatricopeptide repeat (PPR) in ABR17-transgenic A. thaliana, which control physiological, biochemical, and stress signaling cascades due to the imposition of salt stress. Our results are discussed within the context of the effect of the transgene, ABR17, and the roles of miRNA expression may play in mediating plant responses to salinity.





Similar content being viewed by others
References
Accerbi M, Schmidt SA, De Paoli E, Park S, Jeong DH, Green PJ (2010) Methods for isolation of total RNA to recover miRNAs and other small RNAs from diverse species. Methods Mol Biol 592:31–50
Agarwal P, Bhatt V, Singh R, Das M, Sopory SK, Chikara J (2013) Pathogenesis-related gene, JcPR-10a from Jatropha curcas exhibit RNase and antifungal activity. Mol Biotechnol 54:412–425
Arif A, Zafar Y, Arif M, Blumwald E (2012) Improved growth, drought tolerance, and ultrastructural evidence of increased turgidity in tobacco plants overexpressing Arabidopsis Vacuolar Pyrophosphatase (AVP1). Mol Biotechnol 54:379–392
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M et al (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Benes V, Castoldi M (2010) Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 50:244–249
Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G et al (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46:462–476
Bhaskaran S, Savithramma DL (2011) Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot 62:5561–5570
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3:e85
Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J et al (2010) Cell signaling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465:316–321
Carnavale Bottino M, Rosario S, Grativol C, Thiebaut F, Rojas CA, Farrineli L et al (2013) High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS One 8:e59423
Chen Y, Gelfond JA, McManus LM, Shireman PK (2009) Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10:407
Chen L, Zhang Y, Ren Y, Xu J, Zhang Z, Wang Y (2012) Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun 417:892–896
de Lima JC, Loss-Morais G, Margis R (2012) MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet Mol Biol 35:1069–1077
Del Vescovo V, Meier T, Inga A, Denti MA, Borlak JA (2013) Cross-platform comparison of Affymetrix and Agilent microarrays reveals discordant miRNA expression in lung tumors of c-Raf transgenic mice. PLoS One 8:e78870
Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V et al (2012) A PHABULOSA/Cytokinin Feedback Loop Controls Root Growth in Arabidopsis. Curr Biol 22:1699–1704
Derksen H, Rampitsch C, Daayf F (2013) Signaling cross-talk in plant disease resistance. Plant Sci 207:79–87
Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot 103:29–38
Dunfield K, Srivastava S, Shah S, Kav N (2007) Constitutive expression of ABR17 cDNA enhances germination and promotes early flowering in Brassica napus. Plant Sci 173:521–532
Fernandes H, Michalska K, Sikorski M, Jaskolski M (2013) Structural and functional aspects of PR-10 proteins. FEBS J 280:1169–1199
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Gaxiola RA, Fink GR, Hirschi KD (2002) Genetic manipulation of vacuolar proton pumps and transporters. Plant Physiol 129:967–973
Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J et al (2010) Systematic comparsion of microarray profiling, real time PCR and next generation sequencing technologies for measuring differential microRNA expression. RNA 16:991–1006
Gruber V, Blanchet S, Diet A, Zahaf O, Boualem A, Kakar K et al (2009) Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol Genet Genomics 281:55–66
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T (2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol 45:550–559
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
He M, Xu Y, Cao J, Zhu Z, Jiao Y, Wang Y, Guan X et al (2013) Subcellular localization and functional analyses of a PR10 protein gene from Vitis pseudoreticulata in response to Plasmopara viticola infection. Protoplasma 250:129–140
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P et al (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Jung HJ, Kim MK, Kang H (2013) An ABA-regulated putative RNA-binding protein affects seed germination of Arabidopsis under ABA or abiotic stress conditions. J Plant Physiol 170:179–184
Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M et al (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64:1863–1878
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA et al (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S et al (2013) Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS One 8:e52517
Koops P, Pelser S, Ignatz M, Klose C, Marrocco-Selden K, Kretsch T (2011) EDL3 is an F-box protein involved in the regulation of abscisic acid signaling in Arabidopsis thaliana. J Exp Bot 62:5547–5560
Krishnaswamy SS, Srivastava S, Mohammadi M, Rahman MH, Deyholos MK, Kav NN (2008) Transcriptional profiling of pea ABR17 mediated changes in gene expression in Arabidopsis thaliana. BMC Plant Biol 8:91
Krishnaswamy S, Verma S, Rahman MH, Kav NNV (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75:107–127
Kumimoto RW, Siriwardana CL, Gayler KK, Risinger JR, Siefers N, Holt BF 3rd (2013) NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination. PLoS One 8:e59481
Larue CT, Wen J, Walker JC (2009) A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J 58:450–463
Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W (2012) Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol 158:1279–1292
Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, Zheng CC (2013) NFYA1 is involved in regulation of post germination growth arrest under salt stress in Arabidopsis. PLoS One 8:e61289
Liu Q, Feng Y, Zhu Z (2009) Dicer-like (DCL) proteins in plants. Funct Integr Genomics 9:277–286
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Mallory A, Vaucheret H (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22:3879–3889
Mao W, Li Z, Xia X, Li Y, Yu JA (2012) Combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS One 7:e33040
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M et al (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82:113–129
Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M et al (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23:2169–2183
Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P et al (2011) Expression of an Arabidopsis vacuolar H + -pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fiber yield in the field conditions. Plant Biotechnol 9:88–99
Pierre-Jerome E, Moss BL, Nemhauser JL (2013) Tuning the auxin transcriptional response. J Exp Bot 64:2557–2563
Pritchard CC, Cheng HH, Tewari M (2012) microRNA profiling: approaches and considerations. Nature Rev Genet 13:358–369
Sato F, Tsuchiya S, Terasawa K, Tsujimoto G (2009) Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One 4:e5540
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel d (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517-527
Scmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L et al (2008) Real-time PCR quantification of precursor and mature microRNA. Methods 44:31–38
Shuai P, Liang D, Zhang Z, Yin W, Xia X (2013) Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14:233
Srivastava S, Fristensky B, Kav NN (2004) Constitutive expression of a PR10 protein enhances the germination of Brassica napus under saline conditions. Plant Cell Physio 45:1320–1324
Srivastava S, Rahman MH, Shah S, Kav NN (2006) Constitutive expression of the pea ABA-responsive 17 (ABR17) cDNA confers multiple stress tolerance in Arabidopsis thaliana. Plant Biotechnol J 4:529–549
Srivastava S, Emery RJN, Rahman MH, Kav NNV (2007) A crucial role for cytokinins in pea ABR17-mediated enhanced germination and early seedling growth of Arabidopsis thaliana under saline and low temperature stresses. J Plant Growth Reg 26:26–37
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Takeuchi K, Gyohda A, Tominaga M, Kawakatsu M, Hatakeyama A, Ishii N et al (2011) RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol 52:1686–1696
Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Varkonyi-Gasic E, Hellens RP (2011) Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol Biol 744:145–157
Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation are crucial for plant development. Genes Dev 18:1187–1197
Wang L, Dong L, Zhang Y, Zhang Y, Wu W, Deng X et al (2004) Genome-wide analysis of S-Locus F-box-like genes in Arabidopsis thaliana. Plant Mol Biol 56:929–945
Wang B, Howel P, Bruheim S, Ju J, Owen LB, Fodstad O et al (2011) Systematic evaluation of three microRNA profiling platform : microarray, beads array and quantitative real-time PCR array. PLoS One 6:e17167
Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123
Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNAmiR166g and its AtHD-ZIP target genes. Development 132:3657–3668
Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gladiola RA (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H + -pyrophosphatase. Plant Biotechnol J 5:735–745
Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH (2011) starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res (Database issue):D202-229
Yin X, Wang J, Cheng H, Wang X, Yu D (2013) Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237:1213–1225
Zhang X, Liu S, Takano T (2008) Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol 68:131–143
Zhao F, Chen L, Perl A, Chen S, Ma H (2011) Proteomic changes in grape embryogenic callus in response to Agrobacterium tumefaciens-mediated transformation. Plant Sci 181:485–495
Zheng Y, Li YF, Sunkar R, Zhang W (2012) SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res 40:e28
Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H (2010) The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457:1–12
Acknowledgements
Funding from the Natural Sciences and Engineering Research Council (NSERC) of Canada, Agriculture Funding Consortium, Alberta Canola Producers Commission, and Alberta Crop Industry Development Fund is gratefully acknowledged. We would like to thank Sona Todi (Indian Institute of Technology, Kharagpur, India) for her contribution in some of the experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Visualization of the miRNA microarray data through clustering heat maps showing t-test of selected miRNA differentially expressed in transgenic A. thaliana expressing ABR17 gene compared with WT (a), salt stressed ABR17 compared with non-stressed ABR17 (b), salt-stressed WT compared to non-stressed WT (c), and salt-stressed ABR17 as compared to salt-stressed WT (d). Red indicate an increase in abundance, while green represents a decrease in abundance of miRNAs at a P value of less than 0.01 (P < 0.01) (GIF 261 kb)
Supplementary material S2
a-d Excel sheets showing the modulation of expression of miRNAs in transgenic A. thaliana expressing ABR17 gene compared with WT (a), salt stressed ABR17 compared with non-stressed ABR17 (b), salt-stressed WT compared to non-stressed WT (c), and salt-stressed ABR17 as compared to salt-stressed WT (d) (XLS 49 kb)
Rights and permissions
About this article
Cite this article
Verma, S.S., Sinha, R., Rahman, M.H. et al. miRNA-Mediated Posttranscriptional Regulation of Gene Expression in ABR17-Transgenic Arabidopsis thaliana Under Salt Stress. Plant Mol Biol Rep 32, 1203–1218 (2014). https://doi.org/10.1007/s11105-014-0716-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0716-2