Abstract
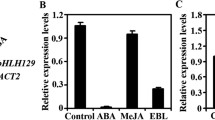
Basic helix-loop-helix (bHLH) transcription factors (TF) comprise a large group of proteins that are involved in many developmental and physiological processes in plants. In this study, a bHLH gene (PkbHLH2), along with its promoter, was cloned from Populus koreana Rehd. A PkbHLH2 promoter::GUS gene fusion construct was generated to investigate the expression of PkbHLH2. The results demonstrated that PkbHLH2 was expressed mainly in leaf stalks, leaf veins and roots. Yeast one-hybrid assays showed that a bZIP gene product (PkbZIP2) can bind specifically to the ABA-responsive elements (ABRE) that exist in the promoter region of PkbHLH2, regulating the expression of PkbHLH2. In addition, the “GC” of the ABRE core motif “ACGTG” was very important for PkbZIP2 recognition, because its mutation to “TT” completely prevented the interaction between PkbZIP2 and ABRE. Furthermore, both PkbHLH2 and PkbZIP2 can be up-regulated by abscisic acid (ABA) and osmotic stress, and share similar expression patterns when exposed to ABA and osmotic stress. These results suggest that PkbZIP2 is an upstream regulator of PkbHLH2, which can control the expression of PkbHLH2 through an ABA-dependent signaling pathway.





Similar content being viewed by others
References
Bai Y, Pattanaik S, Patra B, Werkman J, Xie C, Yuan L (2011) Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234:363–375
Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425–435
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Chinnusamy V, Ohta M, Kanrar S, Lee B, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Feller A, Hernandez JM, Grotewold E (2006) An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J Biol Chem 281:28964–28974
Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116
Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F et al (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23:701–715
Groszmann M, Bylstra Y, Lampugnani ER, Smyth DR (2010) Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J Exp Bot 61:1495–1508
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Heisler M, Atkinson A, Bylstra YH, Walsh R, Smyth DR (2001) SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128:1089–1098
Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H (2010) The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol 51:252–261
Jiang FL, Wang F, Wu Z, Li Y, Shi GJ, Hu JD, Hou XL (2011) Components of the Arabidopsis CBF coold-response pathway are conserved in non-heading Chinese cabbage. Plant Mol Biol Rep 29:525–532
Kim S, Kang J, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE–binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40:75–87
Li H, Sun J, Xu Y, Jiang H, Wu X, Li C (2007) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65:655–665
Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS et al (2008) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228:225–240
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Nieva C, Busk PK, Domínguez-Puigjaner E, Lumbreras V, Testillano PS, Risueño MC, Pagès M (2005) Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Mol Biol 58:899–914
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350
Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15:1998–2006
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W et al (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Rock CD (2000) Tansley review no. 120. Pathways to abscisic acid-regulated gene expression. New Phytol 148:357–396
Tang W, Michael P, Fei YJ, Liu LC, Xu F, Cai XD, Yuan LY, Wu QS, Zhou MQ (2012) Overexpression of AtbZIP60 deltaC gene alleviates salt-induced oxidative damage in transgenic cell cultures. Plant Mol Biol Rep 30:1183–1195. doi:10.1007/s11105-012-0437-3
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770
Xu HM, Wang Y, Chen F, Zhang XZ, Han ZH (2011) Isolation and characterization of the iron-regulated MxbHLH01 gene in Malus xiaojinensis. Plant Mol Biol Rep 29:936–942
Zhang J, Jia W, Yang J, Ismail AM (2006b) Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res 97:111–119
Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J (2011a) The basic helix–loop–helix transcription factor CrMYC2 controls the jasmonate–responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67:61–71
Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM, Kong J, Hua JP (2011b) Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.). Plant Mol Biol Rep 29:626–637
Zhou J, Li F, Wang J, Ma Y, Chong K, Xu Y (2009) Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt-and osmotic stress in Arabidopsis. J Plant Physiol 166:1296–1306
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgment
This work was supported by National Natural Science Foundation of China (31000312).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, L., Gao, C., Wang, Y. et al. A Basic Helix-Loop-Helix Gene from Poplar is Regulated by a Basic Leucine-Zipper Protein and is Involved in the ABA-Dependent Signaling Pathway. Plant Mol Biol Rep 31, 344–351 (2013). https://doi.org/10.1007/s11105-012-0507-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0507-6