Abstract
Background and aims
Arbuscular mycorrhizal fungi (AMF) are beneficial soil microorganisms establishing mutualistic symbioses with most crop plants and promoting plant growth and health. AMF beneficial activities are complemented by their associated microbiota, leading to synergistic interactions positively affecting plant performance. In this work we assessed whether AMF may act as drivers of root bacterial endophytes, facilitating root colonization of host plants by their associated bacteria.
Methods
Two AMF isolates were used, Funneliformis mosseae from Indiana (USA) and Septoglomus sp. from Tuscany (Italy) in an original experimental microcosm system, utilizing micropropagated plants of Prunus persica x Prunus amygdalus inoculated with either intact or mechanically crushed AMF spores, the former able and the latter unable to establish the symbiosis. Spore and root endophytic bacterial communities diversity were analysed by Illumina Miseq sequencing.
Results
This study revealed that AMF with their associated bacteria can shape the root endophytic bacterial communities, inducing differential recruitment depending on the composition of spore-associated microbiota. Such data were consistent between two AMF isolates, associated with diverse bacterial communities, as shown by PERMANOVA, Bray Curtis dissimilarity, hierarchical clustering and indicator species analyses. Moreover, specific bacterial taxa were found exclusively in mycorrhizal roots. Our findings suggested also a differential recruitment depending on the ability of AMF to establish mycorrhizal symbioses.
Conclusion
This work revealed that AMF represent drivers of the endophytic bacterial communities diversity and composition, facilitating root colonization of host plants by their associated bacteria, that become an integral part of the root microbiome as endophytes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants live associated with multifunctional and complex beneficial microbial communities, which establish intimate relationships with their aboveground and belowground tissues. These microorganisms show multiple diverse activities, strongly affecting plant metabolism, nutrition and health (Philippot et al. 2013). Among such microbiota, arbuscular mycorrhizal (AM) fungi (AMF) represent one of the most important group, as they are able to colonize the roots and establish mutualistic symbioses with most crop plants. AMF are obligate biotrophic symbionts and obtain carbon from the plant, in exchange of soil mineral nutrients, absorbed and translocated to the host by the large network of hyphae growing from mycorrhizal roots into the soil environment (Smith and Read 2008). AMF promote plant growth and health and increase plant tolerance to biotic and abiotic stresses, thus reducing the need of chemical fertilizers and pesticides (Bitterlich et al. 2018; El-Sawah et al. 2023; Gianinazzi et al. 2010; Sikes et al. 2009).
AMF beneficial activities cannot be considered separately from the large and diverse bacterial communities living in intimate association with mycorrhizal roots, spores, sporocarps and extraradical hyphae, originating a complex and metabolically active environment called mycorrhizosphere (Faghihinia et al. 2023). Different AMF may harbour diverse bacterial communities, mainly represented by species and genera belonging to Pseudomonadota, Actinobacteriota, Bacillota and Bacteroidota (Agnolucci et al. 2015; Cruz et al. 2008; Emmett et al. 2021; Iffis et al. 2016; Long et al. 2008; Roesti et al. 2005). Several works reported that AMF-associated bacteria showed multifunctional activities as plant growth promoters (PGP) by fixing nitrogen, solubilizing phosphates, mineralizing phytates, producing siderophores and plant hormones (Agnolucci et al. 2019a; Battini et al. 2016; Cruz and Ishii 2011; Sharma et al. 2020) and as mycorrhiza helpers (MH) by promoting spore germination, mycelial growth and symbiosis establishment (Cruz and Ishii 2011; Fernández Bidondo et al. 2016; Sangwan and Prasanna 2022). Overall, such beneficial bacteria may promote and complement AMF functional activities, leading to synergistic interactions positively affecting plant performance (Barea et al. 2002; Giovannini et al. 2020; Turrini et al. 2018). Actually, recent findings demonstrated that P-mobilizing bacteria improve plant growth and P uptake in mycorrhizal wheat, maize and alfalfa (Battini et al. 2017; Wahid et al. 2020; Wang et al. 2023; Zhang et al. 2014).
Interestingly, a recent study revealed that the inoculation of two durum wheat cultivars with Funneliformis mosseae increased the abundance of Actinobacteriota and Bacteroidota inside plant roots. Moreover, it favoured the endophytic establishment of some important PGP genera (Agnolucci et al. 2019b). Another work reported that mycorrhizal symbiosis affected the community composition of endophytic bacteria in lettuce (Han et al. 2023). Alas, these two studies did not investigate the communities of bacteria associated with the mycorrhizal fungus utilised as inoculum. Specific co-inoculation experiments with rhizobia and AMF showed that root colonization and nodule formation by N2-fixing bacteria were facilitated by the presence of AMF (de Novais et al. 2020; Meghvansi et al. 2008; Tajini et al. 2011). These results are noteworthy, as it has long been known that bacterial root endophytes are able to promote plant performance, providing manifold benefits by PGP activities (Hardoim et al. 2015; Liu et al. 2017; Santoyo et al. 2016).
Bacterial endophytes have been found in the roots and stems/leaves of a wide variety of host plants, including important food crops in different ecosystems and geographic areas. The density of endophytic bacteria can reach 104–108 and 103–104 bacterial cells per g of root and stem/leaf tissues, respectively (Hallmann 2001). Communities of bacterial root endophytes are mainly composed by Pseudomonadota, Actinobacteriota, Bacteroidota and Bacillota, but other phyla, such as Acidobacteriota, Chloroflexota, Cyanobacteriota, Planctomycetota, Mycoplasmatota and Verrucomicrobiota may also occur (Ujvári et al. 2021). Root microbiome composition and diversity is influenced by diverse factors, including plant genotype, nutrient status, phenological stage, stress conditions, but also season, farming practices and soil type (Hardoim et al. 2015; Liu et al. 2017).
In this work, we addressed the question as to whether AMF may act as drivers of endophytic root microbiome diversity and composition, facilitating root colonization of host plants by their associated bacteria that could become an integral part of the root microbiome as endophytes. Since different AMF may harbour diverse bacterial communities, the present study was carried out utilizing two AMF isolates, maintained in our culture collection using Cichorium intybus L. and Medicago sativa L. as host plants, differing for their taxonomic and geographic origins: F. mosseae IN101C isolated from Indiana, USA and Septoglomus sp. 14W1, isolated from Tuscany, Italy. To answer the question, a targeted microcosm system was set up, utilizing in vitro-produced micropropagated plants inoculated with either intact or mechanically crushed AMF spores.
Endophytic microbiome composition and diversity of the roots of micropropagated plantlets inoculated with the two AMF was investigated by PCR-based Illumina MiSeq technology, utilized for sequencing the V3-V4 hypervariable region of 16S rRNA gene. This study represents the basis for the management of root beneficial endophytes, AMF and bacteria, whose synergistic activity may provide support for their combined implementation in sustainable agroecosystems.
Materials and methods
Plant and fungal material
Micropropagated plantlets of a selection of Prunus persica x Prunus amygdalus GF677 (hereafter GF677) were obtained from the biotechnological firm Meristema® (Cascine di Buti, Tuscany, Italy). After 5 weeks in the rooting medium, plantlets were removed from the in vitro cultures, their roots were washed in sterile distilled water and inoculated as described below.
The AMF used were two geographically and taxonomically different isolates, maintained in the IMA collection of the Microbiology Laboratories, Department of Agriculture, Food and Environment – DAFE, University of Pisa, Italy: F. mosseae (T.H. Nicolson & Gerd.) C. Walker & A. Schüßler strain IN101C (isolated from a native prairie site near West Lafayette, IN, USA, originally provided by INVAM, Morgantown, WV, USA, and since 1997 maintained in Pisa) and Septoglomus sp. strain 14W1 (isolated from an agricultural field within the UNESCO Man and Biosphere Reserve near Pisa, Tuscany, Italy and maintained in the IMA collection since 2016). The two fungal isolates were grown as pure pot cultures with chicory (Cichorium intybus L.) and alfalfa (Medicago sativa L.) as host plants, and in the same substrate and environmental conditions. Spores (IN101C) and spore clusters (14W1) were extracted from pot culture soil by wet-sieving and decanting, and intact, healthy spores were manually collected with a capillary pipette or forceps under a dissecting microscope (Wild/Leica, Wetzlar, Germany).
Experimental design
For the experiment, 50 intact spores (IN101C) or 15 intact spore clusters (14W1) were placed on sterile tissue paper, which was wrapped around plant roots. These were referenced as belonging to RIND and R14W1 treatments, respectively. The same quantity of spores was mechanically crushed with sterile pestles in Eppendorf tubes and checked under the dissecting microscope for complete disruption, before their utilisation to inoculate additional plantlets, as described above. These plantlets represented treatments RXIND and RX14W1. With the aim of assessing the possible occurrence of environmental root endophytes, replicate uninoculated plantlets were set up in the same way. These are referenced as belonging to treatment RC. Nine replicates for each treatment were used. Plantlets were transferred in 50 mL Falcon tubes containing moist sterile quartz grit. The Falcon tubes were closed in transparent Sun bags (Sigma-Aldrich, St. Louis, MO, USA) and maintained in a growth chamber with 24/21 °C day/night temperatures and 16/8 h light/dark cycle. The experiment was set up in a biological safety cabinet, using sterile tools and materials. Plants were watered as needed and after the second week, 2 mL (once a week) of sterile modified Hoagland’s solution (Hoagland and Arnon 1938) containing ¼ strength of the standard concentration of KH2PO4, were added to each system. After 10 weeks, the plantlets were harvested and the roots were sterilized as indicated by Sun et al. (2008), in order to remove the superficial microbial contaminants. The success of the sterilization process was assessed on 3 × 100 μL of water from the last washing, which were spread-plated on Tryptic Soy Agar (TSA) (Sigma-Aldrich) medium and incubated for 72 h at 28 °C.
For total DNA extraction and subsequent high-throughput sequencing, three biological samples per treatment, each comprised of two pooled root systems, were prepared, in a biological safety cabinet, using sterile tools and stored at -80 °C.
To evaluate mycorrhizal colonization, whole root systems of three replicate plantlets of each treatment were used. The percentage of mycorrhizal root length was evaluated using the grid-line intersect method after root clearing and staining (Giovannetti and Mosse 1980; Turrini et al. 2017).
DNA extraction, amplification and 16S rRNA gene amplicon sequencing
Genomic DNA was extracted from 250 mg of root tissue by grinding with mortar and pestle in liquid nitrogen using the DNeasy® PowerSoil® Pro Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions and eluted in 50 μL 10 mM Tris buffer. In addition, DNA was also extracted from triplicate samples of 50 spores of IN101C and 15 spore clusters of 14W1 used in the experiment (indicated as SIND and S14W1, respectively). The extracted DNA was checked for successful amplification and stored at -20 °C to be used for Illumina MiSeq sequencing. Polymerase chain reaction (PCR) amplification of the hypervariable regions V3-V4 of the 16S rRNA gene was performed using the Pro341F and Pro805R primers (Takahashi et al. 2014). Amplicons were obtained using the Platinum™ Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific, Waltham, MA, USA). Cycle conditions were an initial step at 94 °C (1 min.); 25 cycles of 94 °C (30 s.), 55 °C (30 s.), 68 °C (45 s.); a final extension at 68 °C (7 min.). Libraries were purified using Agencourt AMPure XP (LABPLAN; Naas, Ireland) according to the Illumina metagenomic sequencing library protocol. Dual indices and Illumina sequencing adapters from the Illumina Nextera XT index kits v2 B and C (Illumina, San Diego, USA) were added to the target amplicons in a second index PCR step, according to the Illumina metagenomic sequencing library protocols to generate sequencing index libraries. Sequencing was performed as a 2 × 300 bp paired-end run on the Illumina® MiSeq™ platform. The NGS procedure was performed by BMR Genomics (Padua, Veneto, Italy). Bacterial library preparation and demultiplexing were carried out using the Microbial Ecological tool QIIME2 (Bolyen et al. 2019) version 2021.4.0 pipeline. The high throughput bacterial sequence reads were pre-processed using Cutadapt v.10 (Martin 2011) included in the QIIME2 to eliminate adapter and unwanted primer, followed by denoising, chimeras’ removal, dereplication and OTUs construction using DADA2 (Callahan et al. 2016) at 99% accuracy level. Alignment and taxonomic assignment of bacterial OTUs were done against the Silva (Quast et al. 2012) database version 138. The sequencing data are deposited in the GenBank database with accession numbers PP343280—PP344592.
Statistical analyses
We determined the rarefaction curves to estimate whether the number of screened sequences was sufficient to capture endophytic diversity of each treatment (Supplementary Material Fig. S1). Differences in bacterial endophytes community structures among treatments were assessed by Permutational Multivariate Analysis of Variance (PERMANOVA) on Bray–Curtis distances (nperm = 999). Diversity indices such as Richness (S), Shannon entropy (Hs), Simpson’s dominance (D), were calculated using PAST software (Hammer et al. 2001) version 4.12.
Indicator species analysis (Dufrene and Legendre 1997), was used to identify the endophytic bacterial OTUs indicative of a given inoculation treatment. The indicator value (IndVal) indicated the strength of association, while the statistical significance of association was tested using a permutation test (nperm = 999) at p ≤ 0.05. All the statistical analyses were carried out using PAST version 4.12.
Morpheus-Broad Institute (https://software.broadinstitute.org/morpheus) software was used for the generation of the hierarchical clustering and the heatmap, using average linkage analysis method with one minus Pearson’s correlation as the metric. Root endophytic bacterial communities shared among plants inoculated with intact and crushed spores and uninoculated were defined by Venn diagrams, drawn using BioVenn at https://www.biovenn.nl/index.php.
Results
Bacterial communities of spores and root endophytes
Illumina sequencing produced a total of 383,837 bacterial reads relative to the V3-V4 region of 16S rDNA, which were clustered in 1,382 operational taxonomic units (OTUs) after merging, trimming, and chimera filtering steps. Overall, 1,103 OTUs were grouped in 274 bacterial genera, 169 families, 56 classes, and 20 phyla, mostly represented by Pseudomonadota and Actinobacteriota (63.5%). In addition, 279 OTUs were grouped in 83 taxa which were not identified at the genus level, and which belonged mainly (74.1%) to Pseudomonadota and Actinobacteriota (Fig. S2).
Diversity and composition of bacterial microbiota associated with AMF spores
The reads of bacterial origin obtained from the two spore inocula were 364,740, which clustered in 1,325 OTUs. The number of phyla retrieved in IN101C and 14W1 spores, was 20 and 15, respectively, with about 65% of reads belonging to Pseudomonadota and Actinobacteriota. At the family level, 139 taxa were identified, 123 and 87 in IN101C and 14W1 spores, respectively. The total number of identified genera was 205, comprising 821 OTUs, while other 504 OTUs, unidentified or uncultured, clustered in 138 taxa. The analysis of diversity within each spore community (alpha diversity) showed a richness of 1,051 and 399 OTUs in IN101C and 14W1, respectively, while the richness in identified genera was 181 and 105. Similar dominance levels were found, as shown by Simpson (0.04 vs 0.06) and Shannon (3.7 vs 3.4) indices. The spores of the two AMF shared 125 OTUs (Fig. 1) and 81 identified genera.
The most frequent genera (i.e. those with a frequency higher than 4% of total reads from each spore community) in IN101C spores were Haliangium, Sphingomonas, Nocardioides, Lysobacter, and Bacillus, and in 14W1 Massilia, Lysobacter, Paenarthrobacter, Ramlibacter, Rhizobium group, and Bacillus (Fig. 2).
Many reads (about 32% and 24% of total reads from IN101C and 14W1 spores, respectively) were not identified at the genus level. Most of them belonged to Micrococcaceae, Comamonadaceae, Microscillaceae from both fungal species, to Diplorickettsiaceae, Roseiflexaceae and AKIW781 from IN101C, and to Vicinamibacterales and Oxalobacteriaceae from 14W1. The two microbial communities showed increasing levels of dissimilarity from phylum to OTU, as measured by Bray–Curtis distances. Values of dissimilarities were 0.26 at the phylum level, 0.45 at the family level, 0.67 at the genus level, 0.81 at the OTU level, showing that the bacterial communities associated with the two AMF were very different.
The data obtained showed that the spores of the two AMF isolates were characterised by diverse bacterial communities.
Root endophytic bacterial communities of inoculated plants as compared with those of AMF spores
Overall, the number of reads detected in inoculated roots was 18,580, while in the uninoculated roots only 517 reads were detected, showing that environmental contamination was negligible. The numbers of OTUs retrieved in inoculated roots (105), compared with those occurring in spore bacterial communities, were drastically reduced (5 and 13% in IN101C and 14W1, respectively). Most reads retrieved in inoculated roots (from 51 to 87%) matched with those belonging to the bacterial OTUs retrieved in the spore samples analysed.
The endophytic bacterial communities of the roots inoculated with the two AMF varied significantly at OTU level, as shown by two way PERMANOVA analysis (F = 2.83, P = 0.0004). The multivariate analysis (PERMANOVA) performed on indicator species OTUs (see Fig. 7), confirmed the important role of the two different fungal isolates in shaping the communities (F = 7.93, P = 0.0002).
The data revealed that the two AM symbionts, whose spores were associated with different bacterial communities, significantly affected the diversity and composition of root bacterial endophytes.
Mycorrhizal colonization of roots inoculated with intact or crushed spores
Both IN101C and 14W1 AMF successfully colonized the roots of GF677 plants inoculated with intact spores, with mycorrhizal root length ranging from 25 to 41%. Colonized roots produced large amounts of spores and extraradical mycelium (Fig. 3). No mycorrhizal colonization was detected in the roots of the plants inoculated with crushed spores and in uninoculated plants.
Light photomicrographs of fungal structures formed by Septoglomus sp. 14W1 and Funneliformis mosseae IN101C in the roots of GF677 micropropagated plants 10 weeks after inoculation with intact spores. A, B) Septoglomus sp. 14W1 spores colonizing roots. Scale bars: A) 800 μm; B) 50 μm; C) F. mosseae IN101C spores colonizing roots and extraradical mycelium growing from the roots into the surrounding environment, scale bar: 500 μm; D) appressoria produced by F. mosseae IN101C on the root surface, scale bar: 25 μm. E) Septoglomus sp. 14W1 intraradical hyphae, scale bar: 40 μm; F) Septoglomus sp. 14W1 intracellular arbuscule, scale bar: 20 μm; G) Septoglomus sp. 14W1 entry point, scale bar: 30 μm; H, I) F. mosseae IN101C intraradical hyphae, scale bars: 40 μm and 25 μm, respectively
Root endophytic bacterial communities of plants inoculated with intact or crushed spores
The highest numbers of reads were retrieved in treatments with intact spores, R14W1 and RIND, 7,493 and 5,713, respectively, while treatments with crushed spores, RX14W1 and RXIND, yielded lower numbers, 885 and 4,489, respectively. Overall, 59 and 53 OTUs were detected in roots inoculated with IN101C and 14W1 spores. Among such OTUs, 29 and 24, respectively, matched with those retrieved in the spore samples analysed (Fig. S3).
Exclusive OTUs retrieved in RXIND decreased to 18, compared with 28 found in RIND, while 13 were shared. The decrease was more marked in 14W1 treatments, as exclusive OTUs in RX14W1 were 7, compared with 35 found in R14W1, while 11 were shared (Fig. 4).
Venn diagrams representing OTUs numbers in the roots inoculated with intact and crushed spores of the fungal symbionts Funneliformis mosseae IN101C (A), and Septoglomus sp. 14W1 (B). RXIND and RIND, roots inoculated with F. mosseae IN101C crushed or intact spores, respectively; RX14W1 and R14W1, roots inoculated with Septoglomus sp. 14W1 crushed or intact spores, respectively
The phyla Pseudomonadota and Bacteroidota were well represented (34.7 ± 2.8% and 23.8 ± 5.3%), occurring in all the samples. Actinobacteriota were also well represented (28.4 ± 9.6%), mostly in RXIND (56.5%). Myxococcota, which was the fourth phylum in abundance, was absent in RXIND. Interestingly, Bacillota, largely represented in spore communities, were rare (less than 1% in RIND and RXIND) or absent (in R14W1 and RX14W1).
The analysis of bacterial families showed that in the roots inoculated with both intact and crushed spores of the two AMF, the number of families ranged from 12 to 23, mainly represented by Chitinophagaceae and Micromonosporaceae. Besides, Haliangiaceae was detected in RIND (13.2%) and absent in RXIND, Comamonadaceae represented 18% in mycorrhizal roots and only 0.3% in RXIND. Conversely, Streptomycetaceae was detected in RXIND, where the family represented 25% of the total sequences vs 0.8% in mycorrhizal roots.
Among the IN101C spore-associated bacterial genera, Haliangium, Actinoplanes, Stenotrophomonas, Hyphomicrobium, D05-2 and VHS-B3-70 were exclusively retrieved in mycorrhizal roots inoculated with intact IN101C spores, while among those of 14W1, Streptomyces, Devosia, Desmonostoc_PCC-74, Azospirillum, Piscinibacter, 0319-6G20, Bradyrhizobium and Mycobacterium, were exclusively found in mycorrhizal roots inoculated with intact 14W1 spores.
The main shared genera, detected in the two endophytic communities of RIND and RXIND, were represented by Niastella (22.1% vs 11.7%), Actinoplanes (13.4% vs 5.1%), Rhizobium (7.2% vs 10.3%) and Steroidobacter (4.3% vs 14.1%). The genera characterizing RIND community were Hydrogenophaga (18%) and Haliangium (13.2%), and those characterizing RXIND were Saccharothrix (25.2%) and Streptomyces (25.2%) (Fig. 5). On the other hand, the main shared genera detected in the roots inoculated with intact or crushed 14W1 spores (R14W1 and RX14W1) were represented by Niastella (17.1% vs 29.8%), Haliangium (14.4% vs 11.1%), Ramlibacter (9.5% vs 4.3%), Actinoplanes (7% vs 18.4%), and Steroidobacter (5.7% vs 5.7%). The genera Streptomyces (14%), and 0319-6G20 (7.5%) characterized R14W1 community, while Rhizobium (5.7%) and Variovorax (12.6%) characterized RX14W1 community (Fig. 5).
Distribution of endophytic bacterial genera in the roots of GF677 plants inoculated with intact or crushed spores of the two AMF isolates (Funneliformis mosseae IN101C and Septoglomus sp. 14W1). RXIND and RIND, roots inoculated with F. mosseae IN101C crushed or intact spores, respectively; RX14W1 and R14W1, roots inoculated with Septoglomus sp. 14W1 crushed or intact spores, respectively
Hierarchical clustering analysis showed that bacterial endophytic communities of roots inoculated with IN101C intact spores were different from those inoculated with crushed ones. By contrast, endophytes occurring in the roots inoculated with 14W1 were similar, regardless of the spore inoculum treatment (Fig. 6).
Heatmap and hierarchical clustering analysis summarizing the relative abundance of the bacterial OTUs found in the roots of GF677 plants inoculated with intact or crushed spores of the two AMF isolates (Funneliformis mosseae IN101C and Septoglomus sp. 14W1). RXIND and RIND, roots inoculated with F. mosseae IN101C crushed or intact spores, respectively; RX14W1 and R14W1, roots inoculated with Septoglomus sp. 14W1 crushed or intact spores, respectively. One minus Pearson’s distance was used for clustering. Colours correspond to OTUs’ relative abundance from low (blue) to high (red)
Indicator species analyses of endophytic communities retrieved in the roots of plants inoculated with intact or crushed spores showed characteristic species associated with each different inoculation treatment (P < 0.05) (Fig. 7). In particular, four OTUs (OTU23, OTU44, OTU55, OTU58), which corresponded to Niastella populi (Indval = 82.5), Actinoplanes xinjiangensis (Indval = 50.1), Niastella sp. (Indval = 66.7) and uncultured Hyphomicrobium sp. (Indval = 66.7) were associated with the roots inoculated with intact IN101C spores, while only OTU25, which corresponded to Cupriavidus alkaliphilus (Indval = 66.7) was found strongly associated to the roots of crushed IN101C spores (Fig. 7). Three different OTUs were found associated with roots inoculated with 14W1 intact spores (OTU72, OTU83, OTU89, Indval = 81.7, 66.7, 66.7, respectively) which corresponded to uncultured Ramlibacter sp., Rhizobium sp. and Methylobacillus sp. Three OTUs (OTU45, OTU70, OTU73, Indval = 45.4, 69.2, 60.5, respectively), were found associated with roots inoculated with 14W1 crushed spores (Fig. 7) and corresponded to Haliangium tepidum, Niastella populi and Ohtaekwangia kribbensis.
Significant indicator species of root endophytic communities associated with GF677 roots inoculated with intact or crushed spores of the two AMF isolates (Funneliformis mosseae IN101C and Septoglomus sp. 14W1). RXIND and RIND, roots inoculated with F. mosseae IN101C crushed or intact spores, respectively; RX14W1 and R14W1, roots inoculated with Septoglomus sp. 14W1 crushed or intact spores, respectively. The scale indicates the Indval values. The boxed Indval values are significant at p < 0.05
The diversity of bacterial endophytic communities in roots inoculated with IN101C crushed or intact spores, assessed by Bray Curtis dissimilarity, was 0.84 at the level of OTU, and 0.66 at the level of genera, higher than the diversity of the endophyte communities in roots inoculated with 14W1 spores, which was 0.56 and 0.44, respectively.
Overall, our data showed that the differential enrichment of bacterial endophytes in the roots of plants inoculated with intact or crushed spores (mycorrhizal vs. non-mycorrhizal roots) may be ascribed to the different composition of spore-associated bacterial communities.
Discussion
This study, for the first time, revealed that two AMF isolates, differing for their geographical origin (USA and EU), belonging to different genera and species and associated with diverse bacterial communities, differentially shaped the root endophytic microbiome of the host plants. Our findings suggested also a differential recruitment depending on the ability of AMF to establish the mycorrhizal symbiosis.
Diversity and composition of bacterial microbiota associated with AMF spores
Present data show that the spores of the two AMF isolates were characterised by diverse bacterial communities. The common identified genera were 81, while 24 were exclusively associated to 14W1 and 100 to IN101C, consistently with previous works reporting that spore-associated bacteria differed among diverse AMF isolates. Such differential occurrence may be influenced by specific spore wall composition or spore exudates, supplying nutrients for the nourishment and metabolic activity of the associated microbiota (Agnolucci et al. 2015; Jansa et al. 2013; Roesti et al. 2005; Scheublin et al. 2010; Xu et al. 2023). In particular, large differences in the composition of spore-associated bacteria were found for the genera Massilia and Ramlibacter, abundant in 14W1, and for Sphingomonas and Nocardioides, abundant in IN101C (Fig. 2). Another highly represented, but differentially distributed genus was Paenarthrobacter in 14W1 and IN101C, belonging to the phylum Actinobacteriota, whose members were regularly found in the mycorrhizosphere (Agnolucci et al. 2015; Ames et al. 1989; Filippi et al. 1998). The genus Ramlibacter was previously detected in Gigaspora margarita spore-associated microbiota and Rhizoglomus irregulare (syn. Rhizophagus irregularis) hyphosphere (Long et al. 2008; Wang et al. 2023).
Genera with representation larger than 1%, such as Bacillus, Paenibacillus, Sphingomonas, Rhizobium group, Ensifer, Massilia and Lysobacter, were frequently detected in AMF-associated bacterial communities (Ujvári et al. 2021). Moreover, some bacterial taxa, such as members of the genera Stenotrophomonas, Lysobacter, Bradyrhizobium, Variovorax, Cupriavidus and Bacillus, were previously found capable of utilizing fungal “highways” as means of dispersion (Bravo et al. 2013; de Novais et al. 2020; Pion et al. 2013; Simon et al. 2015).
Spore-associated bacteria affected root endophytic bacterial communities
For the first time, our data, obtained by Illumina sequencing, revealed that the two AM symbionts F. mosseae IN101C and Septoglomus sp 14W1, whose spores were associated with different bacterial communities, significantly affected the diversity and composition of root bacterial endophytes (Fig. 5).
Previous studies reported that the taxonomic composition of root endophytes may be affected by host genotype (Agnolucci et al. 2019b; Walitang et al. 2018; Xu et al. 2020), plant phenological stage (Marques et al. 2015; Van Overbeek and Van Elsas 2008), plant mineral nutrition and agricultural management practices (Hameed et al. 2015; Seghers et al. 2004), soil type and geographic location (Edwards et al. 2015; Samuel et al. 2023; Schlaeppi et al. 2014). Present findings revealed that AMF represent an important microbial source driving the formation and composition of root endophytic bacterial communities.
Mycorrhizal colonization of roots inoculated with intact or crushed spores
The roots of GF677 micropropagated plants inoculated with intact spores showed a good mycorrhizal colonization in plants inoculated with 14W1 and IN101C (Fig. 3). Such colonization levels are consistent with previous data reporting large variability in the colonization ability of diverse AMF species and isolates (Giovannetti et al. 2010; Giovannini et al. 2020; Jansa et al. 2008). Plants inoculated with crushed spores did not establish mycorrhizal colonization, demonstrating the correctness and suitability of our experimental device.
Root endophytic bacterial communities of plants inoculated with intact or crushed spores
The differential enrichment of bacterial endophytes in mycorrhizal roots was mainly ascribed to the different composition of spore-associated bacterial communities. The exclusive or differential occurrence of specific bacterial taxa in mycorrhizal or non-mycorrhizal roots may involve multiple mechanisms, possibly acting simultaneously and/or synergistically. In the case of the inoculation with crushed spores, unable to establish the symbiosis, the associated bacteria adjacent to the roots may behave following a general mechanism, common to many endophytes. Thus, the bacteria are able to colonize the roots after the recognition of host plant exudates and adhesion to the root surface, penetrating either passively through wounds, root fractures, secondary root emergence points, or actively through the action of cell wall–degrading hydrolytic enzymes (Compant et al. 2010). Other factors facilitating root penetration are represented by motility structures and chemical signals aiding chemotaxis (Hardoim et al. 2015; Pinski et al. 2019).
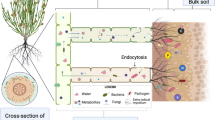
On the other hand, in the case of the inoculation with intact spores, the establishment of the mycorrhizal symbiosis may drive the differential recruitment of bacterial root endophytes in the host plants, as previously suggested by two works, which, alas, neither utilized only spores as inoculum, but mixtures of extraradical mycelium, spores and mycorrhizal root fragments, nor reported the composition of bacterial communities associated with the inoculum (Agnolucci et al. 2019b; Han et al. 2023). Actually, the large communities of bacteria living strictly associated with AMF spores and hyphae may specifically attach and migrate along the hyphae, that may function as highways for bacterial transfer into root tissues during mycorrhizal colonization (de Novais et al. 2020; Jiang et al. 2021; Toljander et al. 2007) (Mechanism 1, Fig. 8). A second mechanism concerns the bacterial microbiota actively interacting with the developing hyphae and their exudates, which may boost the growth of specific taxa or limit the development of others (Cruz and Ishii 2011; Filion et al. 1999; Sharma et al. 2020; Toljander et al. 2007; Xavier and Germida 2003). Such a metabolically active environment may play a role in the selective recruitment of specific bacterial taxa, thus altering the composition of the communities associated with AMF spores and hyphae that could possibly colonize mycorrhizal roots (Mechanism 2, Fig. 8). Moreover, a further mechanism may be active, involving the interaction between fungal symbionts and host plants, which could alter root physiology, exudates and endophytic bacterial microbiomes (Mechanism 3, Fig. 8).
PGP bacteria in root endophytic bacterial communities
The bacterial endophyte sequences obtained in this study were affiliated with bacterial OTUs previously retrieved from the roots of a number of plant taxa, including many important crop species (Adeleke et al. 2021; Ujvári et al. 2021). Most of them have been previously described as PGP bacteria, able to promote plant growth and nutrition. Among the most important, the phylum Actinobacteriota was represented by the genera Streptomyces and Actinoplanes. Isolates of Streptomyces produce several biologically active secondary metabolites and enzymes able to break down insoluble organic polymers, including chitin and cellulose (Seipke et al. 2012), while members of the genus Actinoplanes possess PGP properties, inhibiting plant pathogens and producing a number of compounds, including antifungal compounds, siderophores and hydrolytic enzymes (El-Tarabily 2003; Palaniyandi et al. 2013; Parenti and Coronelli 1979; Vértesy et al. 2000; Wang et al. 2022).
Members of the phylum Bacteroidota were mostly represented by the family Chitinophagaceae. In particular, the genus Niastella reached a consistently high relative abundance in all four root inoculation treatments, accounting for 12–30% of the total bacterial sequences. Niastella spp. were isolated from a wide range of soil and rhizosphere environments (Akter et al. 2021; Weon et al. 2006; Zhang et al. 2010) and detected in root endosphere (Agnolucci et al. 2019b; Dai et al. 2020; Gaggìa et al. 2013), although its potential functional properties are not known. Interestingly, changes in the relative abundance of the genus Haliangium (phylum Myxococcota) in the root endosphere showed a notable enrichment in mycorrhizal roots of IN101C. Haliangium spp. were detected as dominant myxobacterial taxa in soil (Dai et al. 2023; Petters et al. 2021) and in root endosphere (Chu et al. 2021; Dai et al. 2020; Lin et al. 2022). Their functional properties were mainly represented by the production of specific antibiotic substances (haliangicins and haliamide) (Kundim et al. 2003; Sun et al. 2016).
Within the phylum Pseudomonadota (Flores-Félix et al. 2020) the genera Rhizobium, Ensifer and Bradyrhizobium were well represented in all inoculation treatments. Rhizobia are well-known for their beneficial role in plant growth and nutrition, given their ability to fix nitrogen and to act as PGP bacteria (Vargas et al. 2017).
In this study, the family Comamonadaceae was represented by the genera Variovorax and Ramlibacter. The genus Variovorax was found ubiquitously in soil and rhizosphere environments, encompassing metabolically diverse taxa, some known as functional PGP bacteria and/or plant endophytes (Han et al. 2011; Satola et al. 2013) producing ACC-deaminase (Belimov et al. 2009; Chen et al. 2013), IAA (Sun et al. 2018) and siderophores (Natsagdorj et al. 2019). Members of the genus Ramlibacter were reported to degrade cellulose (Kang et al. 2022), which may be the reason behind their endophytic presence.
Conclusions
Two AMF isolates differing for taxonomy, geographical origin and spore-associated bacterial communities differentially modified the root microbiome of the host plants, thus showing that AMF play a key role as drivers of the endophytic bacterial communities colonizing plant roots, representing an important means of transfer of their associated bacteria into plant roots. Data obtained using our original experimental system, utilising intact and mechanically crushed AMF spores as inocula – the former able and the latter unable to establish the symbiosis – suggested that AMF may induce differential recruitment of bacterial root endophytes, depending on their capacity of root colonization, as specific endophytes occurred exclusively in roots inoculated with intact spores.
In the years to come systematic studies on the differential occurrence of root bacterial endophytes in mycorrhizal and non-mycorrhizal plants should be performed, in order to obtain conclusive data on the ability of AMF-associated bacterial communities to establish in the root system as endophytes, and in what ratio.
A further interesting outcome arisen from our work is represented by the fact that most of the endophytic bacterial genera retrieved in mycorrhizal roots and shared with spore-associated bacteria are known as possessing PGP properties. Targeted studies will answer the question as to whether such specific PGP activities, tested in vitro, are maintained when the bacteria become endophytes, thus promoting plant growth, nutrition and health.
This study increased our understanding of the complex network of microbial interactions that may positively affect crop production and represents the basis for the study of AMF and their associated bacteria, whose functional complementarity and synergistic activity might lead to the production of innovative inoculants, with the implementation of beneficial root endophytes for the sustainable intensification of food production systems.
Data availability
The sequencing data are deposited in the GenBank database with accession numbers PP343280—PP344592.
The datasets generated during and/or analysed during the current study are available from the corresponding authors on reasonable request.
References
Adeleke BS, Babalola OO, Glick BR (2021) Plant growth-promoting root-colonizing bacterial endophytes. Rhizosphere 20:100433. https://doi.org/10.1016/j.rhisph.2021.100433
Agnolucci M, Avio L, Pepe A et al (2019a) Bacteria associated with a commercial mycorrhizal inoculum: Community composition and multifunctional activity as assessed by Illumina sequencing and culture-dependent tools. Front Plant Sci 9:1956. https://doi.org/10.3389/fpls.2018.01956
Agnolucci M, Palla M, Cristani C et al (2019b) Beneficial plant microorganisms affect the endophytic bacterial communities of durum wheat roots as detected by different molecular approaches. Front Microbiol 10:2500. https://doi.org/10.3389/fmicb.2019.02500
Agnolucci M, Battini F, Cristani C, Giovannetti M (2015) Diverse bacterial communities are recruited on spores of different arbuscular mycorrhizal fungal isolates. Biol Fertil Soils 51:379–389. https://doi.org/10.1007/s00374-014-0989-5
Akter S, Park JH, Rahman MM, Huq MA (2021) Niastella soli sp. nov., isolated from rhizospheric soil of a persimmon tree. Int J Syst Evol Microbiol 71:004870. https://doi.org/10.1099/ijsem.0.004870
Ames RN, Mihara KL, Bayne HG (1989) Chitin-decomposing actynomycetes associated with a vesicular-arbuscular mycorrhizal fungus from a calcareous soil. New Phytol 111:67–71. https://doi.org/10.1111/j.1469-8137.1989.tb04219.x
Barea JM, Azcón R, Azcón-Aguilar C (2002) Mycorrhizosphere interactions to improve plant fitness and soil quality. Anton Leeuw Int J G 81:343–351
Battini F, Cristani C, Giovannetti M, Agnolucci M (2016) Multifunctionality and diversity of culturable bacterial communities strictly associated with spores of the plant beneficial symbiont Rhizophagus intraradices. Microbiol Res 183:68–79. https://doi.org/10.1016/j.micres.2015.11.012
Battini F, Grønlund M, Agnolucci M et al (2017) Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci Rep 7:4686. https://doi.org/10.1038/s41598-017-04959-0
Belimov AA, Dodd IC, Hontzeas N et al (2009) Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol 181:413–423. https://doi.org/10.1111/j.1469-8137.2008.02657.x
Bitterlich M, Rouphael Y, Graefe J, Franken P (2018) Arbuscular mycorrhizas: A promising component of plant production systems provided favorable conditions for their growth. Front Plant Sci 9:1329. https://doi.org/10.3389/fpls.2018.01329
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bravo D, Cailleau G, Bindschedler S et al (2013) Isolation of oxalotrophic bacteria able to disperse on fungal mycelium. FEMS Microbiol Lett 348:157–166. https://doi.org/10.1111/1574-6968.12287
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Chen L, Dodd IC, Theobald JC et al (2013) The rhizobacterium Variovorax paradoxus 5C–2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J Exp Bot 64:1565–1573. https://doi.org/10.1093/jxb/ert031
Chu C, Fan M, Song C et al (2021) Unveiling endophytic bacterial community structures of different rice cultivars grown in a cadmium-contaminated paddy field. Front Microbiol 12:756327. https://doi.org/10.3389/fmicb.2021.756327
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Cruz AF, Horii S, Ochiai S et al (2008) Isolation and analysis of bacteria associated with spores of Gigaspora margarita. J Appl Microbiol 104:1711–1717. https://doi.org/10.1111/j.1365-2672.2007.03695.x
Cruz AF, Ishii T (2011) Arbuscular mycorrhizal fungal spores host bacteria that affect nutrient biodynamics and biocontrol of soil-borne plant pathogens. Biol Open 1:52–57. https://doi.org/10.1242/bio.2011014
Dai W, Liu Y, Yao D et al (2023) Phylogenetic diversity of stochasticity-dominated predatory myxobacterial community drives multi-nutrient cycling in typical farmland soils. Sci Total Environ 871:161680. https://doi.org/10.1016/j.scitotenv.2023.161680
Dai Y, Yang F, Zhang L et al (2020) Wheat-associated microbiota and their correlation with stripe rust reaction. J Appl Microbiol 128:544–555. https://doi.org/10.1111/jam.14486
de Novais CB, Sbrana C, da Conceição JE et al (2020) Mycorrhizal networks facilitate the colonization of legume roots by a symbiotic nitrogen-fixing bacterium. Mycorrhiza 30:389–396. https://doi.org/10.1007/s00572-020-00948-w
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. https://doi.org/10.1890/0012-9615(1997)067
Edwards J, Johnson C, Santos-Medellín C et al (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112:E911–E920. https://doi.org/10.1073/pnas.1414592112
El-Sawah AM, Abdel-Fattah GG, Holford P et al (2023) Funneliformis constrictum modulates polyamine metabolism to enhance tolerance of Zea mays L., to salinity. Microbiol Res 266:127254. https://doi.org/10.1016/j.micres.2022.127254
El-Tarabily KA (2003) An endophytic chitinase-producing isolate of Actinoplanes missouriensis, with potential for biological control of root rot of lupin caused by Plectosporium tabacinum. Aust J Bot 51:257–266. https://doi.org/10.1071/BT02107
Emmett BD, Lévesque-Tremblay V, Harrison MJ (2021) Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi. ISME J 15:2276–2288. https://doi.org/10.1038/s41396-021-00920-2
Faghihinia M, Jansa J, Halverson LJ, Staddon PL (2023) Hyphosphere microbiome of arbuscular mycorrhizal fungi: a realm of unknowns. Biol Fertil Soils 59:17–34. https://doi.org/10.1007/s00374-022-01683-4
Fernández Bidondo L, Colombo R, Bompadre J et al (2016) Cultivable bacteria associated with infective propagules of arbuscular mycorrhizal fungi. Implications for mycorrhizal activity. Appl Soil Ecol 105:86–90. https://doi.org/10.1016/j.apsoil.2016.04.013
Filion M, St-Arnaud M, Fortin JA (1999) Direct interaction between the arbuscular mycorrhizal fungus Glomus intraradices and different rhizosphere microorganisms. New Phytol 141:525–533. https://doi.org/10.1046/j.1469-8137.1999.00366.x
Filippi C, Bagnoli G, Citernesi AS, Giovannetti M (1998) Ultrastructural spatial distribution of bacteria associated with sporocarps of Glomus mosseae. Symbiosis 24:1–12
Flores-Félix JD, Menéndez E, Peix A et al (2020) History and current taxonomic status of genus Agrobacterium. Syst Appl Microbiol 43:126046. https://doi.org/10.1016/j.syapm.2019.126046
Gaggìa F, Baffoni L, Di Gioia D et al (2013) Inoculation with microorganisms of Lolium perenne L.: evaluation of plant growth parameters and endophytic colonization of roots. New Biotechnol 30:695–704. https://doi.org/10.1016/j.nbt.2013.04.006
Gianinazzi S, Gollotte A, Binet M-N et al (2010) Agroecology: The key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20:519–530. https://doi.org/10.1007/s00572-010-0333-3
Giovannetti M, Avio L, Sbrana C (2010) Fungal spore germination and pre-symbiotic mycelial growth–physiological and genetic aspects. In: Koltai H, Kapulnik Y (eds) Arbuscular mycorrhizas: physiology and function. Springer Science, Dordrecht, pp 3–32
Giovannetti M, Mosse B (1980) An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol 84:489–500
Giovannini L, Palla M, Agnolucci M et al (2020) Arbuscular mycorrhizal fungi and associated microbiota as plant biostimulants: Research strategies for the selection of the best performing inocula. Agronomy 10:106. https://doi.org/10.3390/agronomy10010106
Hallmann J (2001) Plant interactions with endophytic bacteria. In: Jeger MJ, Spence NJ (eds) Biotic Interactions in PlantePathogen Associations. CABI Publishing, Wallingford, United Kingdom, pp 87–119
Hameed A, Yeh MW, Hsieh YT et al (2015) Diversity and functional characterization of bacterial endo- phytes dwelling in various rice (Oryza sativa L.) tissues, and their seed-borne dissemination into rhizosphere under gnotobiotic P-stress. Plant Soil 394:177–197. https://doi.org/10.1007/s11104-015-2506-5
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9 pp, 178kb
Han JI, Choi HK, Lee SW, Orwin PM, Kim J, LaRoe SL et al (2011) Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol 193:1183–1190. https://doi.org/10.1128/jb.00925-10
Han Z, Zhang Z, Li Y et al (2023) Effect of arbuscular mycorrhizal fungi (AMF) inoculation on endophytic bacteria of lettuce. Physiol Mol Plant Pathol 126:102036. https://doi.org/10.1016/j.pmpp.2023.102036
Hardoim PR, van Overbeek LS, Berg G et al (2015) The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. https://doi.org/10.1128/MMBR.00050-14
Hoagland DR, Arnon DI (1938) The water culture methods for growing plants without soil. Circ Calif Agric Exp Stn 347:1–39
Iffis B, St-Arnaud M, Hijri M (2016) Petroleum hydrocarbon contamination, plant identity and arbuscular mycorrhizal fungal (AMF) community determine assemblages of the AMF spore-associated microbes. Environ Microbiol 18:2689–2704. https://doi.org/10.1111/1462-2920.13438
Jansa J, Bukovská P, Gryndler M (2013) Mycorrhizal hyphae as ecological niche for highly specialized hypersymbionts–or just soil free-riders? Front Plant Sci 4:134. https://doi.org/10.3389/fpls.2013.00134
Jansa J, Smith FA, Smith SE (2008) Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol 177:779–789. https://doi.org/10.1111/j.1469-8137.2007.02294.x
Jiang F, Zhang L, Zhou J et al (2021) Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. New Phytol 230:304–315. https://doi.org/10.1111/nph.17081
Kang M, Chhetri G, Kim I et al (2022) Comparative genomic analyses of four novel Ramlibacter species and the cellulose-degrading properties of Ramlibacter cellulosilyticus sp. nov. Sci Rep 12:21233. https://doi.org/10.1038/s41598-022-25718-w
Kundim BA, Itou Y, Sakagami Y et al (2003) New haliangicin isomers, potent antifungal metabolites produced by a marine myxobacterium. J Antibiot 56:630–638. https://doi.org/10.7164/antibiotics.56.630
Lin W, Liu L, Liang J et al (2022) Changes of endophytic microbial community in Rhododendron simsii roots under heat stress and its correlation with leaf physiological indicators. Front Microbiol 13:1006686. https://doi.org/10.3389/fmicb.2022.1006686
Liu H, Carvalhais LC, Crawford M et al (2017) Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552. https://doi.org/10.3389/fmicb.2017.02552
Long L, Zhu H, Yao Q, Ai Y (2008) Analysis of bacterial communities associated with spores of Gigaspora margarita and Gigaspora rosea. Plant Soil 310:1–9. https://doi.org/10.1007/s11104-008-9611-7
Marques JM, da Silva TF, Vollú RE et al (2015) Bacterial endophytes of sweet potato tuberous roots affected by the plant genotype and growth stage. Appl Soil Ecol 96:273–281. https://doi.org/10.1016/j.apsoil.2015.08.020
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Meghvansi MK, Prasad K, Harwani D, Mahna SK (2008) Response of soybean cultivars toward inoculation with three arbuscular mycorrhizal fungi and Bradyrhizobium japonicum in the alluvial soil. Eur J Soil Biol 44:316–323. https://doi.org/10.1016/j.ejsobi.2008.03.003
Natsagdorj O, Sakamoto H, Santiago DMO et al (2019) Variovorax sp. has an optimum cell density to fully function as a plant growth promoter. Microorganisms 7:82. https://doi.org/10.3390/microorganisms7030082
Palaniyandi SA, Yang SH, Zhang L, Suh JW (2013) Effects of actinobacteria on plant disease suppression and growth promotion. Appl Microbiol Biotechnol 97:9621–9636. https://doi.org/10.1007/s00253-013-5206-1
Parenti F, Coronelli C (1979) Members of the genus Actinoplanes and their antibiotics. Annu Rev Microbiol 33:389–411. https://doi.org/10.1146/annurev.mi.33.100179.002133
Petters S, Groß V, Söllinger A et al (2021) The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? ISME J 15:2665–2675. https://doi.org/10.1038/s41396-021-00958-2
Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. https://doi.org/10.1038/nrmicro3109
Pinski A, Betekhtin A, Hupert-Kocurek K, Mur LAJ, Hasterok R (2019) Defining the genetic basis of plant–endophytic bacteria interactions. Int J Mol Sci 20:1947. https://doi.org/10.3390/ijms20081947
Pion M, Bshary R, Bindschedler S et al (2013) Gains of bacterial flagellar motility in a fungal world. Appl Environ Microbiol 79:6862–6867. https://doi.org/10.1128/AEM.01393-13
Quast C, Pruesse E, Yilmaz P et al (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Roesti D, Ineichen K, Braissant O et al (2005) Bacteria associated with spores of the arbuscular mycorrhizal fungi Glomus geosporum and Glomus constrictum. Appl Environ Microbiol 71:6673–6679. https://doi.org/10.1128/AEM.71.11.6673-6679.2005
Samuel SO, Suzuki K, Asiloglu R, Harada N (2023) Rice endophytic communities are strongly dependent on microbial communities specific to each soil. Biol Fertil Soils 59:733–746. https://doi.org/10.1007/s00374-023-01743-3
Sangwan S, Prasanna R (2022) Mycorrhizae helper bacteria: Unlocking their potential as bioenhancers of plant–arbuscular mycorrhizal fungal associations. Microb Ecol 84:1–10. https://doi.org/10.1007/s00248-021-01831-7
Santoyo G, Moreno-Hagelsieb G, del Carmen O-M, Glick BR (2016) Plant growth-promoting bacterial endophytes. Microbiol Res 183:92–99. https://doi.org/10.1016/j.micres.2015.11.008
Satola B, Wübbeler JH, Steinbüchel A (2013) Metabolic characteristics of the species Variovorax paradoxus. Appl Microbiol Biotechnol 97:541–560. https://doi.org/10.1007/s00253-012-4585-z
Scheublin TR, Sanders IR, Keel C, Van Der Meer JR (2010) Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J 4:752–763. https://doi.org/10.1038/ismej.2010.5
Schlaeppi K, Dombrowski N, Oter RG, Themaat VLVE, Schulze-Lefert P (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA 111:585–592. https://doi.org/10.1073/pnas.1321597111
Seghers D, Wittebolle L, Top EM, Verstraete W, Siciliano SD (2004) Impact of agricultural practices on the Zea mays L. endophytic community. Appl Environ Microbiol 70:1475–1482. https://doi.org/10.1128/AEM.70.3.1475-1482.2004
Seipke RF, Kaltenpoth M, Hutchings MI (2012) Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev 36:862–876. https://doi.org/10.1111/j.1574-6976.2011.00313.x
Sharma S, Compant S, Ballhausen M-B et al (2020) The interaction between Rhizoglomus irregulare and hyphae attached phosphate solubilizing bacteria increases plant biomass of Solanum lycopersicum. Microbiol Res 240:126556. https://doi.org/10.1016/j.micres.2020.126556
Sikes BA, Klironomos CK, JN, (2009) Plant and fungal identity determines pathogen protection of plant roots by arbuscular mycorrhizas. J Ecol 97:1274–1280. https://doi.org/10.1111/j.1365-2745.2009.01557.x
Simon A, Bindschedler S, Job D et al (2015) Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol Ecol 91:fiv116. https://doi.org/10.1093/femsec/fiv116
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Academic Press, London
Sun L, Qiu F, Zhang X et al (2008) Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol 55:415–424. https://doi.org/10.1007/s00248-007-9287-1
Sun Y, Tomura T, Sato J et al (2016) Isolation and biosynthetic analysis of haliamide, a new PKS-NRPS hybrid metabolite from the marine myxobacterium Haliangium ochraceum. Molecules 21:59. https://doi.org/10.3390/molecules21010059
Sun SL, Yang WL, Fang WW et al (2018) The plant growth-promoting rhizobacterium Variovorax boronicumulans CGMCC 4969 regulates the level of indole-3-acetic acid synthesized from indole-3-acetonitrile. Appl Environ Microbiol 84:e00298–e00318. https://doi.org/10.1128/AEM.00298-18
Tajini F, Trabelsi M, Drevon J-J (2011) Co-inoculation with Glomus intraradices and Rhizobium tropici CIAT899 increases P use efficiency for N2 fixation in the common bean (Phaseolus vulgaris L.) under P deficiency in hydroaeroponic culture. Symbiosis 53:123–129. https://doi.org/10.1007/s13199-011-0117-3
Takahashi S, Tomita J, Nishioka K et al (2014) Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 9:e105592. https://doi.org/10.1371/journal.pone.0105592
Toljander JF, Lindahl BD, Paul LR et al (2007) Influence of arbuscular mycorrhizal mycelial exudates on soil bacterial growth and community structure. FEMS Microbiol Ecol 61:295–304. https://doi.org/10.1111/j.1574-6941.2007.00337.x
Turrini A, Agnolucci M, Palla M et al (2017) Species diversity and community composition of native arbuscular mycorrhizal fungi in apple roots are affected by site and orchard management. Appl Soil Ecol 116:42–54. https://doi.org/10.1016/j.apsoil.2017.03.016
Turrini A, Avio L, Giovannetti M, Agnolucci M (2018) Functional complementarity of arbuscular mycorrhizal fungi and associated microbiota: The challenge of translational research. Front Plant Sci 9:1407. https://doi.org/10.3389/fpls.2018.01407
Ujvári G, Turrini A, Avio L, Agnolucci M (2021) Possible role of arbuscular mycorrhizal fungi and associated bacteria in the recruitment of endophytic bacterial communities by plant roots. Mycorrhiza 31:527–544. https://doi.org/10.1007/s00572-021-01040-7
Van Overbeek L, Van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol 64:283–296. https://doi.org/10.1111/j.1574-6941.2008.00469.x
Vargas LK, Volpiano CG, Lisboa BB et al (2017) Potential of rhizobia as plant growth-promoting rhizobacteria. In: Zaidi A, Khan MS, Musarrat J (eds) Microbes for legume improvement, 2nd edn. Springer Int Publ, New York, pp 153–174
Vértesy L, Ehlers E, Kogler H et al (2000) Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. II. Isolation and Structural Characterization J Antibiot 53:816–827. https://doi.org/10.7164/antibiotics.53.816
Wahid F, Fahad S, Danish S et al (2020) Sustainable management with mycorrhizae and phosphate solubilizing bacteria for enhanced phosphorus uptake in calcareous soils. Agriculture 10:334. https://doi.org/10.3390/agriculture10080334
Walitang DI, Kim CG, Kim K et al (2018) The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biol 18:51. https://doi.org/10.1186/s12870-018-1261-1
Wang P, Yang L, Sun J et al (2022) Structure and function of rhizosphere soil and root endophytic microbial communities associated with root rot of Panax notoginseng. Front Plant Sci 12:752683
Wang L, Zhang L, George TS, Feng G (2023) A core microbiome in the hyphosphere of arbuscular mycorrhizal fungi has functional significance in organic phosphorus mineralization. New Phytol 238:859–873. https://doi.org/10.1111/nph.18642
Weon HY, Kim BY, Yoo SH et al (2006) Niastella koreensis gen. nov., sp. nov. and Niastella yeongjuensis sp. nov., novel members of the phylum Bacteroidetes, isolated from soil cultivated with Korean ginseng. Int J Syst Evol Microbiol 56:1777–1782. https://doi.org/10.1099/ijs.0.64242-0
Xavier LJC, Germida JJ (2003) Bacteria associated with Glomus clarum spores influence mycorrhizal activity. Soil Biol Biochem 35:471–478. https://doi.org/10.1016/S0038-0717(03)00003-8
Xu Y, Chen Z, Li X, T et al (2023) Mycorrhizal fungi alter root exudation to cultivate a beneficial microbiome for plant growth. Funct Ecol 37:664–675. https://doi.org/10.1111/1365-2435.14249
Xu Y, Ge Y, Song J, Rensing C (2020) Assembly of root-associated microbial community of typical rice cultivars in different soil types. Biol Fertil Soils 56:249–260. https://doi.org/10.1007/s00374-019-01406-2
Zhang L, Fan J, Ding X et al (2014) Hyphosphere interactions between an arbuscular mycorrhizal fungus and a phosphate solubilizing bacterium promote phytate mineralization in soil. Soil Biol Biochem 74:177–183. https://doi.org/10.1016/j.soilbio.2014.03.004
Zhang K, Wang Y, Tang Y et al (2010) Niastella populi sp. nov., isolated from soil of Euphrates poplar (Populus euphratica) forest, and emended description of the genus Niastella. Int J Syst Evol Microbiol 60:542–545. https://doi.org/10.1099/ijs.0.012112-0
Funding
Open access funding provided by Università di Pisa within the CRUI-CARE Agreement. This study received financial support from the University of Pisa (Fondi di Ateneo).
Author information
Authors and Affiliations
Contributions
Monica Agnolucci, Luciano Avio, Manuela Giovannetti and Alessandra Turrini conceived, planned and designed the experiments; Caterina Cristani, Arianna Grassi, Irene Pagliarani, Gergely Ujvàri and Alessandra Turrini carried out the experiments; Alessandra Turrini and Gergely Ujvàri carried out mycorrhizal colonization assessment and molecular and data analyses; Manuela Giovannetti, Alessandra Turrini and Gergely Ujvàri wrote the first draft of the manuscript and all authors commented on previous versions of the manuscript. This paper was part of GU’s doctoral thesis work at the University of Pisa. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Anna-Maria Pirttilä.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gergely Ujvári and Arianna Grassi These two authors are designed as co-first authors
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ujvári, G., Grassi, A., Avio, L. et al. Root endophytic bacterial communities are shaped by the specific microbiota associated to mycorrhizal symbionts. Plant Soil (2024). https://doi.org/10.1007/s11104-024-06801-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11104-024-06801-9