Abstract
Purpose
Ureides, allantoin and allantoate, are N-rich compounds used for N transport in nodulated ureide legumes. Here, we investigated their role in response of Phaseolus vulgaris and Pisum sativum, representing ureide and amide legumes, respectively, to Cd toxicity.
Methods
First, ureide content and ureide metabolism in P. vulgaris and P. sativum grown under control conditions and treated with 50 μM CdCl2 for 48 hours was investigated. Then, the effect of exogenous allantoin and its precursor, uric acid, on Cd-related oxidative lesion was examined in both legumes.
Results
Cd increased the content of both ureides only in the leaves of P. vulgaris, which was consistent with transcript levels and activity of ureide metabolic enzymes, and was accompanied by an increase in uric acid content. In P. sativum leaves, Cd increased the activity of ureide biosynthesis enzymes and decreased the activity of ureide degradation enzymes, although the uric acid content did not change, while the allantoin and allantoate contents were significantly reduced. Exogenous uric acid and allantoin suppressed Cd-induced H2O2 and O2 accumulation and alleviated the effects of oxidative damage measured by RNA degradation, chlorophyll and malondialdehyde content in both legumes.
Conclusions
P. sativum use allantoin and uric acid as antioxidant agents to mitigate Cd-related oxidative tissue damage. In P. vulgaris, the involvement of the ureide pathway in Cd-induced N salvage and recycling is rather a priority.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most of the species belonging to the Fabaceae family are able to fix atmospheric nitrogen through symbiosis with soil bacteria known as rhizobia (Andrews and Andrews 2017). This gives them an advantage over nonlegumes when grown in nitrogen-poor soil conditions and can reduce the need for nitrogen fertilization. The N fixed in the nodules of legumes can be exported to the shoot via the xylem mainly in the form of ureides (allantoin, allantoate) or amides (asparagine, glutamine); therefore, a distinction can be made between ureide (e.g., Phaseolus vulgaris, Glycine max) and amide legumes (e.g., Pisum sativum, Vicia faba) (Schubert 1986).
Ureides are N-rich molecules with an N:C ratio of 1:1, which enables them to provide fixed N to sink tissues at minimal C cost. They are formed by the oxidative catabolism of purines (Baral et al. 2016; Todd et al. 2006; Werner and Witte 2011; Witte and Herde 2020; Zrenner et al. 2006). The first common intermediate in the degradation pathway of all purine bases is xanthine, which is hydroxylated in the cytosol by xanthine dehydrogenase (XDH; EC 1.17.1.4) to hypoxanthine and then to uric acid (Montalbini 2000). Subsequently, uric acid is oxidized in peroxisomes by urate oxidase (UO; EC 1.7.3.3) to 5-hydroxyisourate, which is then hydrolyzed and decarboxylated to allantoin by the bifunctional transthyretin-like protein (Lamberto et al. 2010; Pessoa et al. 2010; Werner and Witte 2011). Further ureide degradation occurs in the ER via four enzymatic steps, releasing four NH4+, two CO2, and one glyoxylate from one ureide molecule. Allantoin is first metabolized to allantoate by allantoin amidohydrolase (allantoinase; ALN; EC 3.5.2.5) (Duran and Todd 2012; Raso et al. 2007), which is followed by hydrolysis of allantoate to (S)-ureidoglycine by allantoate amidohydrolase (AAH; E.C. 3.5.3.9) (Díaz-Leal et al. 2014; Todd and Polacco 2006; Werner et al. 2008). Next, (S)-ureidoglycine is converted to (S)-ureidoglycolate by ureidoglycine aminohydrolase (EC 3.5.3.26) (Serventi et al. 2010), and finally, ureidoglycolate amidohydrolase (EC 3.5.1.116) hydrolyzes (S)-ureidoglycolate to glyoxylate (Wells and Lees 1991; Werner et al. 2013).
The purine catabolic pathway has also been described in nonlegumes, where it has been primarily associated with remobilization and recycling of nitrogen, particularly during senescence (Brychkova et al. 2008; Soltabayeva et al. 2018) and seedling development (Quiles et al. 2009). This pathway may also play a pivotal role in plant responses to adverse environmental conditions. However, most of the stress-related studies were conducted on Arabidopsis, where increased accumulation of ureides, mainly allantoin, was observed under salinity (Irani and Todd 2016), sulfate deficiency (Nikiforova et al. 2005), nitrogen deficiency (Soltabayeva et al. 2018), drought (Irani and Todd 2016), NH4+ treatment (Lescano et al. 2020), high irradiance (Irani et al. 2018), prolonged darkness (Brychkova et al. 2008), Cd stress (Nourimand and Todd 2016), or UV-C irradiation and wounding (Soltabayeva et al. 2022). In Arabidopsis, allantoin accumulation is regulated mainly at the transcriptional level by induction of XDH1, UO expression and down-regulation of ALN and AAH (Brychkova et al. 2008; Irani and Todd 2016; Irani et al. 2018; Lescano et al. 2016; Nourimand and Todd 2016; Soltabayeva et al. 2022).
In legumes, ureide metabolism in response to stress factors was investigated predominantly under drought (Alamillo et al. 2010; Coleto et al. 2014; Díaz-Leal et al. 2014). Higher levels of allantoate were observed in the leaves and roots of drought-stressed P. vulgaris in both nonnodulated plants fed NO3− and plants cultivated under N2 fixation conditions. This suggests that under adverse environmental conditions, ureide accumulation may be independent of N fixation and may be a general plant stress response (Alamillo et al. 2010; Coleto et al. 2014). In P. vulgaris, strong induction of ALN expression and posttranscriptional inhibition of AAH have been proposed as major causes of allantoate accumulation under drought conditions (Alamillo et al. 2010; Coleto et al. 2014; Díaz-Leal et al. 2014).
The role of ureides under stress conditions is still not fully understood, although attempts to explain it have been the subject of several studies, mainly in Arabidopsis. Aln mutants with increased allantoin content showed higher tolerance to osmotic stress, salinity, drought, and Cd treatments, which was associated with lower levels of reactive oxygen species (ROS) (Irani and Todd 2016; Nourimand and Todd 2016; Watanabe et al. 2014) and increased production of abscisic acid (ABA) (Watanabe et al. 2014). While the reactivity of ureides against ROS was shown to be limited in rice seeds (Wang et al. 2012), allantoin applied to E. coli (Gus’kov et al. 2002) as well as allantoin and allantoate applied to Arabidopsis leaves has been shown to act as ROS scavengers (Brychkova et al. 2008). Additionally, allantoin was shown to activate the antioxidant enzymes, superoxide dismutase and ascorbate peroxidase, in both control and Cd-treated Arabidopsis plants (Nourimand and Todd 2016), whereas in cucumber plants, allantoin-mediated attenuation of Cd toxicity was due to increased accumulation of ascorbate and glutathione (Dresler et al. 2019).
Cadmium (Cd) is one of the most abundant metals in a contaminated environment and is easily taken up by plants, leading to many harmful symptoms. Cd damages the photosynthetic apparatus and suppresses stomatal opening, thus inhibiting photosynthesis and transpiration (Liu et al. 2011) and interfering the uptake and movement of water and mineral nutrients (Nazar et al. 2012). It also triggers oxidative stress, possibly by disrupting the electron transport chain, impairing antioxidant defenses, or increasing NADPH oxidase activity (Cuypers et al. 2010). Therefore, several defense mechanisms have evolved in plants to mitigate Cd toxicity. For example, plants utilize the ROS detoxification machinery, which includes both antioxidant enzymes (e.g., ascorbate peroxidase, catalase, superoxide dismutase, glutathione reductase) and non-enzymatic scavengers such as ascorbate, glutathione, or carotenoids (Cuypers et al. 2010). To survive Cd toxicity, plants also synthesize additional protective compounds, such as proline or phytochelatins, as well as signaling molecules, such as ABA, salicylic acid or nitric oxide (Stroiński et al. 2013; Zdunek-Zastocka et al. 2021). In the presented work, we hypothesized that ureides may also be part of plant defense mechanisms against Cd toxicity, at least in legumes.
In order to confirm the above hypothesis, the effect of Cd treatment on the content of allantoin and allantoic acid in leaves and roots of Phaseolus vulgaris and Pisum sativum, representing ureide and amide legumes, respectively, was first investigated. Additionally, to understand the mechanisms underlying the Cd-induced changes in the content of the two ureides, biochemical and molecular approaches were employed to study the metabolism of ureides and the possible role of allantoin and its precursor, uric acid, in alleviating Cd-induced oxidative stress in both legumes.
Materials and methods
Plant material and growth conditions
Seeds of Phaseolus vulgaris (cv. Saxa) and Pisum sativum (cv. Iłówiecki) were germinated in moist vermiculite. Twelve-day-old seedlings were transferred to containers filled with aerated 1/2 Hoagland medium (Hoagland and Arnon 1938). Nitrate was applied as 4 mM NaNO3. After two days, the medium was supplemented with CdCl2 (50 μM), allantoin (200 μM), uric acid (200 μM) or a combination thereof. Plants grown in medium without CdCl2 served as controls. After 48 h of treatments, the plants (twelve plants per treatment) were harvested and separated into roots and leaves. For both species, analyzes were performed on fully developed leaves (Fig. S1). Samples collected from twelve plants were combined, frozen in liquid N2 immediately after harvest and stored at −80 °C until use.
Experiments were conducted in a plant growth chamber (the MLR-352H Climate Chamber, PHC Biomedical – formerly Panasonic Biomedical) at a humidity of 60% and a light intensity of 300 μmol s−1 m−2. The air temperature was 22 °C (16 h) during the day and 19 °C (8 h) at night. The experiment was repeated independently four times, giving four independent biological replicates.
Quantification of ureides and uric acid
For determination of ureides, plant samples were ground in 50 mM K-phosphate buffer, pH 7.0, at a ratio1:8 (w/v), then incubated at 100 °C for 20 min and centrifuged at 16000 g and 4 °C for 20 min (Takagi et al. 2018; Lescano et al. 2020). Quantification of ureides was performed in the resulting supernatants by applying the differential conversion of ureide compounds to glyoxylate and colorimetric detection of glyoxylate derivatives as described by Vogels and van der Drift (1970). Allantoin, allantoate and total ureide concentrations were calculated according to Lescano et al. (2020).
Uric acid was extracted in a 1:1 (v/v) mixture of 30 mM Na-citrate and 55 mM Na-acetate, pH 5.2 (Corpas et al. 1997). The homogenate was centrifuged at 16000 g and 4 °C for 20 min, ultrafiltrated with UltraFree centrifuge filters (10 kDa, Millipore), then passed through a 0.45 μm syringe-filters (Millipore). Uric acid was then determined by a high-performance liquid chromatography using a Shimadzu HPLC system, model LC-2050C 3D-Prominence-i equipped with a photodiode array detector (PDA) and a Platinum C18-EPS column, 5 μm, 250 × 4.6 mm (Alltech Associates). The mobile phase was a 1:1 (v/v) mixture of 30 mM Na-citrate and 55 mM Na-acetate, pH 5.2 for leaves extracts and pH 4.7 for roots extracts. The flow rate was 0.5 ml × min−1, the sample injection volume was 20 μl, the column temperature was 30 °C, and the total run time was 26 min. The detection of the uric acid was performed at 289 nm and was based on its retention time, absorption spectrum and coelution with standard added to the samples to be analyzed. The amount of uric acid was calculated from the peak area and the linear regression equation of the calibration curve.
Determination of uricase activity
Extraction and partial purification of uricase was performed according to Capote-Mainez and Sánchez (1997) with some modifications. Plant tissue (1 g) was homogenized in 4 ml of extraction buffer (100 mM K-phosphate buffer, pH 8.0, 50 mM KCl, 5 mM MgCl2, 2 mM EDTA, 2 mM DTT, 4% polyvinylpolypyrrolidone) using a prechilled mortar and pestle. The extracts were centrifuged at 18000 g and 4 °C for 30 min, and the supernatants were collected and again centrifuged as above. Proteins in the resulting supernatants were precipitated with 60% acetone at −20 °C for 2 h, centrifuged at 5000 g and 4 °C for 20 min, resuspended in 2 ml of extraction buffer, and dialyzed overnight at 4 °C against 20 mM K-phosphate buffer, pH 8.0, 7 mM β-ME, and 2 mM EDTA. The enzyme solutions were centrifuged at 16000 g and 4 °C for 30 min, and the supernatants were recovered and used to measure UO activity as described previously by Bergmann et al. (1983). The standard assay mixture contained 780 μl of 50 mM Tricine-KOH buffer, pH 8.0, 200 μl of the partially purified enzyme solution, and 20 μl of 6.25 mM uric acid (final 0.125 mM). The decrease in urate content due to oxidation was measured at 293 nm.
Determination of allantoinase activity
Plant tissues were ground in 1:3 (w/v) 50 mM Tricine-NaOH, pH 8.0, containing 2 mM MnSO4 and 0.15% (w/v) Na-deoxycholate. The extracts were then centrifuged twice at 16000 g and 4 °C for 20 min, and the resulting supernatants were used for ALN activity assay as described by Duran and Todd (2012). The standard reaction mixture was composed of 20 μl of plant extract, 180 μl of extraction buffer and 200 μl of 20 mM allantoin. The enzymatic reaction was carried out at 30 °C for 30 min and was terminated by the addition of 200 μl of 0.15 M HCl. Allantoate was finally determined spectrophotometrically as a glyoxylate derivative at 535 nm (Vogels and Van der Drift 1970).
Determination of allantoate amidohydrolase activity
Plant tissues were ground in 1:2 (w/v) 50 mM Tricine-NaOH, pH 8.0, containing 2 mM MnSO4, 150 mM NaCl, 15 mM DTT, 0.5% Triton X-100, and 50 μM phenyl phosphorodiamidate (Díaz-Leal et al. 2014). The extracts were then centrifuged twice at 16000 g and 4 °C for 20 min and passed through a Sephadex G-25 desalting column (0.5 × 4.5 cm; GE Healthcare) equilibrated with the extraction buffer. The protein-containing fractions were pooled and used for AAH activity assay as described by Werner et al. (2008) and used by Díaz-Leal et al. (2014). Briefly, hydrolysis of allantoate was performed in a reaction mixture containing 150 μl of dialyzed plant extracts, 250 μl of 6 mM allantoate and 100 μl of the extraction buffer at 37 °C, which was followed by heating at 100 °C for 10 min. The ammonia concentration was then estimated by the Nessler method as described by Molins-Legua et al. (2006).
Xanthine dehydrogenase activity staining
Plant tissues were ground using a prechilled mortar and pestle in 250 mM Tris–HCl, pH 8.5, containing 1 mM EDTA, 10 mM glutathione and 2 mM dithiothreitol, at a ratio of 1:3 (w/v) (Zdunek-Zastocka and Lips 2003a). The extracts were then centrifuged twice at 16000 g and 4 °C for 20 min, and the resulting supernatants were subjected to native polyacrylamide gel electrophoresis (PAGE) on 7.5% polyacrylamide gels (Laemmli 1970). Gel wells were loaded with 50 μg of root proteins or 200 μg of leaf proteins. After electrophoresis, gels were stained in 0.1 M Tris-HCl, pH 7.5, containing 0.1 mM PMS (phenazine methosulphate), 1 mM MTT (3(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), and 1 mM xanthine at 25 °C in the dark. The relative activity of XDH was calculated based on the intensity of the resulting formazan bands using ImageJ software (http://imagej.nih.gov/ij). The intensity of the formazan bands is directly proportional to the enzyme activity (Rothe 1974; Coleto et al. 2019).
Total RNA extraction and cDNA synthesis
Total RNA was isolated according to the method described by Chomczynski and Sacchi (1987), except that RNA was precipitated on ice by adding 0.3 volume of 10 M LiCl. The isolated RNA was treated with DNase I, and a total of 2 μg of RNA was reverse transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s instructions.
Identification of nucleotide sequences of genes involved in ureide metabolism in P. sativum
Internal fragments of the analyzed genes (PsXDH, PsALN, PsAAH) were amplified using Q5 high-fidelity DNA Polymerase (New England BioLabs) and primers designed from known sequences of Medicago truncatula XDH (XM_003597388), ALN (XM_003593472), AAH (XM_013607669). The primers are listed in Table S1. PCR conditions were as follows: 30 s at 98 °C; 40 cycles of 10 s at 98 °C, 30 s at 57 °C, and 1.5 min (PsALN, PsAAH) or 3 min (PsXDH) at 72 °C; followed by a final extension for 10 min at 72 °C. PCR products were cloned into the pJET1.2 vector (Thermo Scientific), amplified in E. coli JM109 and sequenced in the DNA Sequencing and Oligonucleotide Synthesis Laboratory at The Institute of Biochemistry and Biophysics, Polish Academy of Sciences.
Gene expression analysis
Real-time PCR was performed using a LightCycler® 96 instrument (Roche). The reaction mixture contained 5 μl of SYBR Green I Master Mix (Roche), 3 μl of cDNA template (equivalent to 10 ng of total RNA), and 0.5 μM of each primer in a total volume of 10 μl. Thermal cycling conditions were as follows: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, 15 s at 63 °C, and 20 s at 72 °C. Melting curves (95 °C for 10 s, 65 °C for 1 min, and 97 °C for 1 s in continuous acquisition mode) were generated for each reaction to ensure amplification specificity. The relative expression levels of target genes were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). Genes encoding β-tubulin and actin were used as internal controls (Die et al. 2010). All primers used for real-time PCR are listed in Tables S2 and S3. All reactions were performed at least in triplicate for each of the three biological replicates.
Determination of the content of reactive oxygen species
Superoxide radical (O2−) content was measured by monitoring the nitrite formation from hydroxylamine in the presence of O2− as described by Elstner and Heupel (1976) and used by others (Jung et al. 2020; Zhang et al. 2022), with some modifications. Briefly, the plant tissues were ground in 65 mM K-phosphate buffer, pH 7.8, containing 7.5 mM hydroxylamine hydrochloride, at a ratio of 1:4 (w/v). The homogenates were centrifuged at 13000 g and 4 °C for 15 min, and 0.1 ml of the resulting supernatants were mixed with 0.17 ml of the extraction buffer and 0.03 ml of 50 mM hydroxylamine hydrochloride, and incubated at 25 °C for 20 min. Then, 0.3 ml of 58 mM sulfanilamide and 0.3 ml of 7 mM N-(1-naphthyl)ethylenediamine were added, and the mixture was again incubated at 25 °C for 20 min. Finally, the same volume of chloroform was added, mixed and the reaction mixture was centrifuged at 13000 g for 5 min. The absorbance of upper phase was measured at 530 nm.
H2O2 content was determined as described by Jana and Choudhuri (1981). Plant tissues were ground in 50 mM K-phosphate buffer, pH 6.5, at a ratio of 1:10 (w/v). The homogenate was centrifuged at 6000 g for 25 min. Then, 0.6 ml of the supernatant was mixed with 0.2 ml of 0.1% titanium chloride (in 20% H2SO4), and the mixture was centrifuged at 6000 g for 15 min. The absorbance of the supernatant was measured at 410 nm.
Determination of the content of chlorophyll and malondialdehyde
Total chlorophyll was extracted and analyzed using dimethyl sulfoxide (DMSO) as described by Wellburn (1994). Briefly, leaves (50 mg) were ground in liquid N2, vortexed with 2 ml of dimethyl sulfoxide (DMSO) and incubated at 65 °C for 60 min. The residues were removed by centrifugation at 13000 g for 10 min, and the absorbance of the extracts was measured at 649 and 665 nm against DMSO as a blank.
Malondialdehyde (MDA) content was measured according to the method described by Heath and Packer (1968). Plant tissue (150 mg) was homogenized with 1.5 ml of 0.1 M K-phosphate buffer, pH 6.8. The homogenates were centrifuged at 13000 g for 15 min. The supernatants were mixed 1:1 with TBA (0.5% thiobarbituric acid in 15% trichloroacetic acid) and incubated at 100 °C for 20 min. After cooling, the reaction mixtures were centrifuged at 13000 g for 10 min, and the absorbance of the supernatants was measured at 532 nm and 600 nm.
Statistical analysis
Results are the means of four independent experiments (biological replicates). Analytical determinations and enzymatic assays were performed three times (technical replicates) for each biological experiment. The data were statistically analyzed using one-way analysis of variance (ANOVA) for Cd effect and two-way analysis of variance (ANOVA) for simultaneous Cd and ureide effect, separately in leaves and roots. The conformity of data with a normal distribution was verified using the Shapiro-Wilk test, and the homogeneity of variances across the data sets was validated using Levene’s test. Then, to assess the significance of differences between group means, parametric Tukey’s test was used for data with the normal distribution and equal variances (the vast majority of analyses) or non-parametric Kruskal-Wallis test for data that are normally distributed but with unequal variances (effect of Cd on PvUO expression in leaves of P. vulgaris, effect of Cd on O2− content in roots of P. vulgaris in the group of plants grown with uric acid). Statistical analysis was performed using the Statistica ver. 9.1 software.
Results
Ureide content and metabolism in Phaseolus vulgaris as affected by Cd
To investigate whether Cd induces changes in the content and metabolism of ureides in legumes, the content of allantoin and allantoate and the activity of enzymes engaged in their synthesis and degradation were studied in P. vulgaris and P. sativum, representing ureide and amide legumes, respectively, which were exposed for 48 h to 50 μM CdCl2.
As a result of Cd treatment, an increase in the content of the two analyzed ureides was observed only in the leaves of P. vulgaris (Fig. 1). In the leaves of P. vulgaris, the contents of allantoin and allantoate were almost 4 times higher in the Cd-treated plants than in the controls. In leaves, Cd treatment also increased the activity of UO, ALN and AAH, yet lower increase was noticed in AAH activity (Fig. 2). Notably, the accumulation of a particular ureide resulted, from a higher rate of its synthesis than the rate of its processing further along the pathway. For example in plants exposed to Cd, UO activity was 3-fold higher than in controls, whereas the activity of ALN activity was only twice as high and of AAH even less (30%), resulting in an increase in both allantoin and allantoate content. In roots, the content of allantoin and allantoate decreased by about 45% in response to the Cd treatment, as did the activities of UO, ALN and AAH.
Allantoin and allantoate content in leaves and roots of Phaseolus vulgaris (a) and Pisum sativum (b) exposed to Cd treatment for 48 h. Cadmium was applied as 50 μM CdCl2. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means of control and Cd treated plants, separately for roots and leaves, are shown above the columns by different letters
Activity of uricase (a, b), allantoinase (c, d) and allantoate amidohydrolase (e, f) in leaves (a, c, e) and roots (b, d, f) of Phaseolus vulgaris (a, c, e) and Pisum sativum (b, d, f) exposed to Cd treatment for 48 h. Cadmium was applied as 50 μM CdCl2. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means of control and Cd treated plants, separately for leaves and roots, are shown above the columns by different letters
Ureide content and metabolism in Pisum sativum as affected by Cd
In P. sativum, treatment with cadmium decreased the total ureide content in both leaves and roots (Fig. 1b). In leaves, the Cd-induced decrease in ureide content was mainly due to decrease in the content of allantoic acid (by about 80%) and to a lesser extent of allantoin (by about 30%). In roots, Cd treatment decreased the content of allantoin and allantoate by about 60% and 40%, respectively. In Cd-treated plants, UO activity was higher in leaves (by 20%) and similar in roots compared to that of control plants, while the activity of ALN was similar in leaves and lower in roots (by 25%) compared to that of control plants, and AAH activity was lower than in controls, in both leaves (by 15%) and roots (by 25%) (Fig. 2).
Since the ureide content in the expanded leaves of P. sativum did not increase after Cd treatment, as would be expected from the analysis of the activity of the enzymes involved in ureide metabolism, the ureide content in the young not fully expanded leaves were also examined (Fig. S2). There was a presumption that under Cd stress, ureides might move from older to younger leaves and accumulate in the later leaves. For comparison, the content of ureides in response to Cd in the young not fully expanded trifoliate leaves of P. vulgaris was also examined (Fig. S2). The content of ureides in the younger leaves of P. sativum was twice as high as in the older leaves, yet Cd treatment decreased the total ureide content in both type of leaves, regardless of their stage of development (Figs. S2b, Fig. 1b). For comparison, Cd treatment increased the content of both ureides in young leaves of P. vulgaris about 3–5 times (Fig. S2a), as did in unifoliate leaves (Fig. 1a).
Transcript levels of enzymes involved in ureide metabolism as affected by Cd
To understand the molecular mechanisms underlying the Cd-induced changes in ureide content, real-time PCR analyzes for UO, ALN, and AAH were performed for both leaves and roots of P. vulgaris and P. sativum after 48 h of growth on media containing 50 μM CdCl2.
Since not all nucleotide sequences of the genes encoding enzymes of ureide metabolism in P. sativum were available in gene databases, it was necessary to identify at least fragments of the coding sequences of these genes. The first identified nucleotide sequence fragment (3176 bp) shares a high percent identity with the sequences of XDH genes from Arabidopsis thaliana (AtXDH1, NM_119655; AtXDH2, NM_119656; 73%) (Hesberg et al. 2004), Brassica rapa (BrXDH, XM_009109780, 73%) (Yi et al. 2021), Medicago truncatula (MtXDH, XM_003597388, 91%), Phaseolus vulgaris (PvXDH, XM_007150309, 87%) (Coleto et al. 2019), or Vitis vinifera (VvXDH, XM_002285437, 78%) (You et al. 2017) and was designated as Pisum sativum XDH (PsXDH, OL441840) (Fig. S3). The second identified nucleotide sequence (1359 bp) shows a high percentage of identity with the sequences of ALN genes from Arabidopsis thaliana (AtALN, NM_116734, 71%) (Irani and Todd 2016; Werner et al. 2008), Glycine max (GmALN1, Glyma15g07910, 85%; GmALN2, Glyma13g31430, 85%; GmALN3, Glyma15g07920, 86%; GmALN4, Glyma13g31420, 87%) (Duran and Todd 2012), Medicago truncatula (MtALN, XM_003593472, 91%), and Phaseolus vulgaris (PvALN1, XM_007148116, 84%; PvALN2, XM_007148115, 85%) (Alamillo et al. 2010; Duran and Todd 2012) and was designated as Pisum sativum ALN (PsALN, OL441838) (Fig. S4). The third identified nucleotide sequence fragment (1102 bp) shares a high percent identity with the sequences of AAH genes from Arabidopsis thaliana (AtAAH, NM_118126, 74%) (Werner et al. 2008), Glycine max (GmAAH, NM_001349411, 89%) (Charlson et al. 2009; Werner et al. 2008), Medicago truncatula (MtAAH, XM_013607669, 94%), or Phaseolus vulgaris (PvAAH, EF650088, 88%) (Díaz-Leal et al. 2014) and was designated as Pisum sativum AAH (PsAAH, OL441839) (Fig. S5).
The mRNA levels of UO and ALN increased after Cd treatment in the leaves of both legumes, by approximately 250 and 125%, respectively, in P. vulgaris and by 100 and 70%, respectively, in P. sativum (Fig. 3). The Cd-induced up-regulation of UO and ALN expression in leaves corresponded to an increase in UO and ALN activity in P. vulgaris (Fig. 2), whereas in P. sativum there is most likely a posttranscriptional down-regulation of UO and ALN activities (Fig. 2). In roots, the expression of UO and ALN decreased by approximately 50% in P. vulgaris and by 30% in P. sativum under Cd stress (Fig. 3), corresponding to a decrease in enzymatic activities in both legumes (Fig. 2). Expression of AAH was not significantly altered by Cd treatment in either leaves or roots of P. vulgaris or P. sativum (Fig. 3), suggesting posttranscriptional regulation of AAH in both organs of both legumes.
Transcript levels of genes encoding uricase (a, b), allantoinase (c, d) and allantoate amidohydrolase (e, f) in leaves and roots of Phaseolus vulgaris (a, c, e) and Pisum sativum (b, d, f) exposed to Cd treatment for 48 h. Cadmium was applied as 50 μM CdCl2. Transcripts levels of target genes in Cd-stressed plants were expressed relative to those of control plants, which were set to 1, after normalization to reference genes (β-tubulin and actin). Primers used for PCR analyses are listed in Table S2 and S3. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means of control and Cd treated plants, separately for leaves and roots, are shown above the columns by different letters
Activity and transcript level of XDH and content of uric acid as affected by Cd
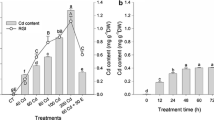
To clarify whether the Cd-induced changes in ureide content are related to the changes in the level of their precursor, the content of uric acid as well as the expression and activity of XDH in the leaves and roots of P. vulgaris and P. sativum were examined after 48 h of growth on media containing 50 μM CdCl2.
As a result of Cd treatment, XDH activity increased by 120 and 90% in both P. vulgaris and P. sativum, respectively, but only in leaves (Fig. 4). In contrast, XDH activity in the roots of both legumes was about 30% lower in Cd-treated plants than in controls (Fig. 4a, b). Like activity, expression of XDH was influenced by Cd in both legumes. In leaves, transcript levels of PvXDH and PsXDH were 100% higher in Cd-treated plants than in controls, while in roots by 50% (Fig. 4c, d). Thus, the Cd-induced increase in PvXDH and PsXDH expression observed in leaves of both legumes was accompanied by an increase in XDH activity, whereas the Cd-induced posttranscriptional down-regulation of XDH activity probably occurs in roots.
Activity of xanthine dehydrogenase (a, b), transcript level of XDH (c, d), and content of uric acid (e, f) in leaves and roots of Phaseolus vulgaris (a, c, e) and Pisum sativum (b, d, f) exposed to 50 μM CdCl2 for 48 h. The numbers above the zymograms indicate relative values obtained by density scanning and analysis by ImageJ software. Zymograms are representative of similar results obtained for four independent biological replicates. Transcript levels of XDH in Cd-stressed plants were expressed relative to those of control plants, which were set to 1, after normalization to reference genes (β-tubulin and actin). Primers used for PCR analyses are listed in Table S2.The results in the graphs are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means of control and Cd treated plants, separately for leaves and roots, are shown above the columns by different letters
In the leaves, application of Cd significantly increased the uric acid content in P. vulgaris (by about 200%), but not in P. sativum, whereas in the roots, uric acid content was increased by about 40% in both legumes (Fig. 4e, f).
Exogenously applied allantoin and uric acid enhance resistance of legumes to Cd toxicity
The results described above show that neither uric acid or allantoin accumulation was observed in leaves of P. sativum treated with Cd, although an increase in the rate of their synthesis and a decrease in the rate of their breakdown was noticed. Therefore, it was further investigated whether the observed status quo could be due, at least in part, to the use of these compounds in the alleviation of Cd-induced oxidative stress, whether in leaves or roots. Accordingly the effect of uric acid (200 μM) and allantoin (200 μM), which were applied to the growth medium in parallel with 50 μM CdCl2, on the production H2O2 and O2− and the level of other stress markers in leaves and roots of P. sativum and P. vulgaris was examined after 48 h of treatment.
The production of ROS enhanced significantly after Cd treatment, as a 2–3-fold increase in H2O2 concentration (Fig. 5a, b) and a 3–4-fold increase in O2− content were observed in the leaves and roots of both legumes (Fig. 5c, d). The application of uric acid almost completely removed an excess of ROS resulting from Cd treatment, while the application of allantoin managed to remove 80 to 95% of ROS enhancement, suggesting contribution of both ureides to limit ROS production (Fig. 5).
Changes in H2O2 (a, b) and O2− content (c, d) in leaves and roots of Phaseolus vulgaris (a, c) and Pisum sativum (b, d) exposed to allantoin or uric acid for 48 h. Allantoin (All) and uric acid (UA) were applied to the growth medium at a concentration of 200 μM, simultaneously with 50 μM CdCl2. Plants grown without CdCl2 were designates as controls. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means are shown above the columns by different letters
Under Cd stress, biomass production [leaf and root dry matter (DW)] was significantly reduced, as a 25 and 35% decrease in DW per plant was noticed for Phaseolus vulgaris and Pisum sativum, respectively (Figs. 6a and 7a). Simultaneous application of uric acid almost completely abolished the Cd-induced biomass reduction in both legumes, while application of allantoin attenuated the Cd-mediated DW reduction about 50%.
Changes in dry weight (a), chlorophyll content (b), RNA content (c), and malondialdehyde content (d) in Phaseolus vulgaris exposed to allantoin or uric acid for 48 h. Allantoin (All) and uric acid (UA) were applied to the growth medium at a concentration of 200 μM, simultaneously with 50 μM CdCl2. Plants grown without CdCl2 were designates as controls. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means are shown above the columns by different letters
Changes in dry weight (a), chlorophyll content (b), RNA content (c), and malondialdehyde content (d) in Pisum sativum exposed to allantoin or uric acid for 48 h. Allantoin (All) and uric acid (UA) were applied to the growth media at a concentration of 200 μM, simultaneously with 50 μM CdCl2. Plants grown without CdCl2 were designates as controls. The results are the means (± SD) of four biological replicates (n = 4). Significant differences (at least p < 0.05) between the means are shown above the columns by different letters
Cd toxicity and associated oxidative stress caused also a decrease in chlorophyll content (about 25%) and an increase in the content of the lipid peroxidation resulting product, MDA (by 35–55%), a marker that reflects the degree of membrane damage induced by stressors such as Cd (Figs. 6b, d, 7b, d). Additionally, Cd induced the significant degradation of RNA (by 35–50%), which occurred to a similar extend in both legumes (Figs. 6c, 7c). Notably, exogenously applied uric acid and allantoin attenuated the Cd-induced decrease of chlorophyll content (about 95% by uric acid, and 70% by allantoin) and attenuated the increase of MDA content (about 85–95% by uric acid, and 70–80% by allantoin), and also reversed Cd-induced degradation of RNA (about 80–90% by uric acid, and about 60–75% by allantoin).
Discussion
Plants are constantly exposed to a wide range of environmental stimuli that have detrimental effects on their growth and productivity. To minimize the adverse effects of unfavorable environmental conditions, plants have developed various defense mechanisms (Cuypers et al. 2010; Stroiński et al. 2013; Zdunek-Zastocka et al. 2021). It has been suggested that allantoin and allantoic acid may also be part of these mechanisms, as increases in ureide content have been observed in plant responses to salinity (Irani and Todd 2016; Sagi et al. 1998), dehydration (Alamillo et al. 2010; Coleto et al. 2014) or wounding (Soltabayeva et al. 2022). In our study, it was found that also 50 μM CdCl2 applied for 48 h induces the accumulation allantoin and allantoic acid, however, only in leaves of P. vulgaris, but not in P. sativum (Fig. 1), even though the degree of Cd-induced oxidative stress was similar in both legumes, as estimated by changes in the content of chlorophyll, H2O2 and MDA, a product of lipid peroxidation (Figs. 5, 6, 7). When lower concentrations of Cd (1, 2.5 and 5 μM CdCl2) were applied to the nutrient medium for a longer period (7 days), similarly the increase of both ureides was observed only in the leaves of P. vulgaris (Fig. S6). It is well known that under nodulation conditions, ureides, purine degradation products, are transported from the roots of tropical legumes through the xylem to the shoot (Schubert 1986). Given the decrease of allantoin and allantoate content in the roots of Cd-treated legumes (Fig. 1), the translocation of ureides from roots to leaves could also be considered as a potential mechanism for enriching the ureide pool observed in leaves of Cd-treated P. vulgaris. However, in P. vulgaris roots, Cd treatment suppressed the activity of XDH, a catalytic bottleneck in purine catabolism, as well as UO, a following enzyme of the ureide biosynthesis from purines (Figs. 2 and 4). This may suggest that the contribution of root-derived ureides in their accumulation observed in leaves under Cd stress may be lower than under nodulation conditions, when the rate of ureide biosynthesis from purines in roots is significantly increased (Baral et al. 2016). As mentioned above, under Cd treatment, accumulation of both allantoin and allantoic acid takes place in P. vulgaris leaves regardless of their stage of development (Fig. 1, S2, S6). It has been shown that the metabolism of ureides in the unifoliate leaves of Cd-treated P. vulgaris favors a higher rate of ureide synthesis and a lower rate of ureide degradation (Figs. 2 and 4), which enables their local accumulation. However, it cannot be excluded that some of ureides of unifoliate origin may move to younger trifoliate leaves. This is the case in Arabidopsis plants, where purine degradation products provide an additional source of N remobilized from older to younger leaves under N-deficient conditions, which was supported by increased expression of ureide permeases, key components of ureide transport observed in older leaves (Soltabayeva et al. 2018).
So far, Cd-induced increase in ureide content was reported only for Arabidopsis leaves and involved only allantoin (Nourimand and Todd 2016). In Arabidopsis leaves, Cd increased allantoin content 1.5-fold, which was associated with increased expression of AtUO, and decreased transcript levels and enzymatic activity of ALN (Nourimand and Todd 2016). In P. vulgaris leaves, Cd treatment increased not only UO activity, but also the activity of ALN and AAH (Fig. 2), with the increase in the activity of the enzyme synthesizing the respective ureide always higher than that of the enzyme degrading it, leading to the accumulation of the respective ureide. An increase in the ureide content, but mainly only of allantoate, was previously observed in P. vulgaris leaves in response to drought, under both nonfixing and N2-fixing conditions (Alamillo et al. 2010). It was concluded that the accumulation of ureides in tissues other than nodules is likely part of a general response to stress and is highly uncoupled from nitrogen fixation (Alamillo et al. 2010). In this study, a Cd-induced increase in allantoin and allantoic acid in the leaves of P. vulgaris was also observed under non-N-fixing conditions and may support the above assumption. Increased accumulation of allantoate in drought-stressed P. vulgaris has been associated with a strong induction of ALN expression (Alamillo et al. 2010; Coleto et al. 2014) and, to a lesser extent, with posttranscriptional inhibition of AAH (Charlson et al. 2009; Díaz-Leal et al. 2014). Increased concentrations of allantoin and allantoic acid in the leaves of Cd-stressed P. vulgaris were associated with higher transcription and activity of enzymes responsible for both production (UO) and degradation (ALN) of allantoin (Figs. 2 and 3). In addition, Cd treatment increased the activity of AAH, an enzyme that catalyzes allantoate hydrolysis; however, the PvAAH transcript levels were not affected. Our results suggest that Cd-induced changes in ureide metabolism observed in leaves of P. vulgaris are regulated at the transcriptional level by induction of XDH, UO and ALN gene expression, and posttranscriptionally by upregulation of AAH. Such regulation in both transcriptional and posttranscriptional manner should result in the accumulation of ureides, mostly allantoate but also allantoin (Fig. 1).
Purines used for ureide biogenesis can be generated not only by de novo synthesis but also by turnover of nucleic acids (Baral et al. 2016). Therefore it is possible that under Cd stress, ureides are important for recycling and recovery of N from Cd-induced degradation of nucleic acids. The degradation of nucleic acids associated with ureide accumulation has been described during natural or dark-induced leaf senescence (Brychkova et al. 2008; Lambert et al. 2017; Soltabayeva et al. 2018) and was also observed in Cd-treated legumes in the presented study (Figs. 6c and 7c). Thus, in P. vulgaris treated with Cd, where increase of the ureide content and of activities of XDH, UO, ALN and AAH was observed at both the activity and transcript level (except of PvAAH), the ureide pathway may be involved in Cd-induced N recycling.
In P. sativum leaves, treatment of plants with 50 μM Cd decreased the content of allantoin and allantoic acid, regardless of the source and douse of N, both in leaves and roots (Fig. 1). In leaves, the observed decrease in allantoin content occurred even though the rate of allantoin production slightly increased (Fig. 2), whereas the rate of allantoin degradation decreased (Fig. 2). Because uric acid content did not change significantly under Cd stress in the leaves, and even increased in the roots (Fig. 4e, f), the Cd-induced decrease in allantoin content was more likely unrelated to the severe shortage of its precursor. Moreover, in the leaves Cd significantly induced the activity and transcript level of XDH (Fig. 4a-d), the enzyme that converts hypoxanthine and xanthine to uric acid. Therefore, the question arose as to why we did not observe an increase in the levels of uric acid and allantoin in the leaves, since the analysis of the activity of enzymes involved in their metabolism suggests that this is the case. One way to explain this would be that both uric acid and allantoin are used as antioxidant agents, thus protecting the plants from oxidative damage. As mentioned earlier, increased production of ROS in plant cells is an early symptom of Cd toxicity leading to oxidation of lipids, proteins and nucleic acids (Cuypers et al. 2010). Therefore, the higher XDH activity observed in leaves of Cd-treated P. sativum as well as P. vulgaris (Fig. 4a, b) may be due to the fact that uric acid production is necessary to support the ROS scavenging machinery (Sun et al. 2021; Ma et al. 2016). Indeed, an increase in XDH activity has previously been observed under unfavorable environmental stimuli such as salinity, drought, an excess of NH4+ ions or prolonged darkness (Brychkova et al. 2008; Watanabe et al. 2010; Zdunek-Zastocka and Lips 2003b).
Although chloroplasts and mitochondria have been considered the most important sources of ROS, in recent years attention has also been paid to peroxisomes as one of the main contributors to the production of ROS in cells (Corpas et al. 2020). Peroxisomes have an large battery of H2O2-generating enzymes, for example glycolate oxidase, acyl-CoA oxidase, urate oxidase, polyamine oxidase, sulfite oxidase, sarcosine oxidase, superoxide dismutase, which are involved in multiple biochemical pathways like photorespiration, fatty acid beta-oxidation or the catabolism of polyamines or nucleic acids. Peroxisomes contain also xanthine oxidase, which generates superoxide radical, and nitric oxide synthase which can produce nitric oxide. In addition to the subcellular compartments mentioned above, ROS are also produced in the cytosol, where are formed notably through the activity of NADPH oxidase, aldehyde oxidase, xanthine dehydrogenase, and nitric oxide synthase (Forrester et al. 2018). To control the steady-state levels of these reactive molecules, a complex set of ROS scavenging systems, including both enzymatic and non-enzymatic ROS scavengers, operates in individual subcellular compartments (Janků et al. 2019). Uric acid produced in the cytosol by XDH and in peroxisomes by XO, as well as allantoin, which is synthetized in peroxisomes, may be another elements of this antioxidant defense machinery, neutralizing excessive amounts of ROS produced under stress conditions.
Uric acid is known to be a potent antioxidant in land animals and may be responsible for up to 55% of the antioxidant capacity to scavenge free radicals in human plasma at physiologically appropriate concentrations (Ames et al. 1981; Jakše et al. 2019). In Arabidopsis, urate infiltration into the leaves was effective in reducing peroxynitrite-related oxidative damage (Alamillo and García-Olmedo 2001), while the addition of exogenous uric acid to the MS medium prevented the accumulation of H2O2 in the chloroplasts of the xdh1 mutant (Ma et al. 2016). Thus, uric acid can act as an effective scavenger also in plants, alleviating the oxidative tissue damage caused by multiple environmental stress factors. Indeed, pretreatment with exogenous uric acid attenuated the drought-hypersensitive phenotype of XDH-suppressed Arabidopsis lines and increased the resistance of apples to salt stress (Watanabe et al. 2010; Sun et al. 2021). In our study, the application of uric acid almost completely removes the excess of ROS caused by Cd treatment and alleviated the symptoms of oxidative damage as measured by biomass production, RNA degradation, chlorophyll and MDA content in both P. vulgaris and P. sativum (Figs. 5, 6 and 7), supporting the engagement of uric acid in the neutralization of ROS in plants.
Both endogenous and exogenous allantoin has been shown to protect Arabidopsis leaves from darkness-induced oxidative damage to chlorophyll as well as protecting leaves from exogenously applied H2O2 (Brychkova et al. 2008) and to reduce oxidative stress under drought or salinity (Irani and Todd 2016), suggesting its possible role in ROS scavenging. Although allantoin has been found to alleviate Cd-induced adverse symptoms by activating antioxidant enzymes or accumulating non-enzymatic radical scavengers, respectively, in Arabidopsis and cucumber plants (Nourimand and Todd 2016; Dresler et al. 2019), its direct reactivity against ROS still requires further investigation and explanation (Gus’kov et al. 2002; Wang et al. 2012). In our study, exogenous application of allantoin significantly attenuated Cd-induced oxidative stress in both P. vulgaris and P. sativum, albeit lower than exogenously applied uric acid (Figs. 5, 6 and 7). Although allantoin accumulation was observed only in P. vulgaris leaves and not in P. sativum (Fig. 1a, b), yet, since in P. sativum we observed an increased rate of allantoin synthesis and a reduced rate of its degradation (Fig. 2), a role of allantoin in direct ROS scavenging in P. sativum exposed to Cd cannot be ignored.
The results presented in this study show the different responses of P. vulgaris and P. sativum, representing amide and ureide legumes, respectively, to Cd treatment in terms of ureide content and metabolism, especially in leaves. These differences are primarily due to post-translational modifications of enzymes involved in ureide metabolism. It was observed that Cd affects the expression of ureide metabolism genes in a similar manner in P. vulgaris and P. sativum, in both leaves and roots (Fig. 3), but the activities of UO, ALN, AAH in response to Cd differ significantly between the two species, especially in leaves (Fig. 2). While Cd strongly induced the expression of UO and ALN in leaves, a significant increase in the activity of both enzymes was observed only in P. vulgaris, whereas in P. sativum the activity of UO changed only slightly and the activity of ALN decreased. Differences were also found in the activity of AAH, which was induced by Cd in P. vulgaris leaves and reduced in P. sativum (Fig. 2), although transcript levels were not significantly affected by Cd in both species (Fig. 3). The differences in ureide content and metabolism between ureide and amide legumes were observed previously in the early stage of seedling development, when ureide content and allantoinase activity in cotyledons and embryonic axes was higher in ureide than amide plants (Quiles et al. 2019). in addition, superoxide dismutase, guaiacol peroxidase, and ascorbate peroxidase activities in both embryonic axes and cotyledons differed significantly among these legume groups, suggesting that ureidic and amidic plants may employ different strategies to cope with the oxidative stress associated with the development of seedlings, for example (Quiles et al. 2019).
In summary, the ureide and amide legumes, represented here by P.vulgaris and P. sativum respectively, use ureides and the ureide pathway in different way to mitigate the deleterious effects of Cd toxicity. Amide legumes could use allantoin and its precursor, uric acid, as antioxidant agents to mitigate Cd-induced oxidative tissue damage. Ureide legumes, on the other hand, could use the ureide pathway not only for protecting from Cd-related oxidative stress, but also to recycle N from Cd-induced degradation of nucleic acids.
Data availability
Data supporting the findings of this study are available in the supplementary material of this article.
Abbreviations
- AAH:
-
Allantoate amidohydrolase
- ALN:
-
Allantoinase
- UO:
-
Urate oxidase
- XDH:
-
Xanthine dehydrogenase
References
Alamillo JM, García-Olmedo F (2001) Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J 25:529–540. https://doi.org/10.1046/j.1365-313x.2001.00984.x
Alamillo JM, Diaz-Leal JL, Sanchez-Moran MV, Pineda M (2010) Molecular analysis of ureide accumulation under drought stress in Phaseolus vulgaris L. Plant Cell Environ 33:1828–1837. https://doi.org/10.1111/j.1365-3040.2010.02187.x
Ames BN, Cathcart R, Schwiers E, Hochstein P (1981) Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci U S A 78:6858–6862. https://doi.org/10.1073/pnas.78.11.6858
Andrews M, Andrews ME (2017) Specificity in legume-rhizobia symbioses. Int J Mol Sci 18:705. https://doi.org/10.3390/ijms18040705
Baral B, Teixeira da Silva JA, Izaguirre-Mayora ML (2016) Early signaling, synthesis, transport and metabolism of ureides. J Plant Physiol 193:97–109. https://doi.org/10.1016/j.jplph.2016.01.013
Bergmann H, Preddie E, Verma DPS (1983) Nodulin-35: a subunit of specific uricase (uricase II) induced and localized in the uninfected cells of soybean nodules. EMBO J 12:2333–2339. https://doi.org/10.1002/j.1460-2075.1983.tb01743.x
Brychkova G, Fluhr R, Sagi M (2008) Formation of xanthine and the use of purine metabolites as a nitrogen source in Arabidopsis plants. Plant Signal Behav 3:999–1001. https://doi.org/10.4161/psb.6304
Capote-Mainez N, Sánchez F (1997) Characterization of the common bean uricase II and its expression in organs other than nodules. Plant Physiol 115:1307–1317. https://doi.org/10.1104/pp.115.4.1307
Charlson DV, Korth KL, Purcell LC (2009) Allantoate amidohydrolase transcript expression is independent of drought tolerance in soybean. J Exp Bot 60:847–851. https://doi.org/10.1093/jxb/ern332
Chomczynski P, Sacchi N (1987) Single-step method of total RNA isolation by a single extraction with an acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. https://doi.org/10.1006/abio.1987.9999
Coleto I, Pineda M, Rodino AP, De Ron AM, Alamillo JM (2014) Comparison of inhibition of N2 fixation and ureide accumulation under water deficit in four common bean genotypes of contrasting drought tolerance. Ann Bot 113:1071–1082. https://doi.org/10.1093/aob/mcu029
Coleto I, Pineda M, Alamillo JM (2019) Molecular and biochemical analysis of XDH from Phaseolus vulgaris suggest that uric acid protects the enzyme against the inhibitory effects of nitric oxide in nodules. Plant Physiol Biochem 143:364–374. https://doi.org/10.1016/j.plaphy.2019.09.008
Corpas FJ, Colina CDL, Sanchez-Rasero F, Rio LAD (1997) A role for leaf peroxisomes in the catabolism of purines. J Plant Physiol 151:246–250. https://doi.org/10.1016/S0176-1617(97)80161-7
Corpas FJ, González-Gordo S, Palma JM (2020) Plant peroxisomes: a factory of reactive species. Front Plant Sci 11:853. https://doi.org/10.3389/fpls.2020.00853
Cuypers A, Plusquin M, Remans T, Jozefczak M, Keunen E, Gielen H, Opdenakker K, Nair AR, Munters E, Artois TJ, Nawrot T, Vangronsveld J, Smeets K (2010) Cadmium stress: an oxidative challenge. Biometals 23:927–940. https://doi.org/10.1007/s10534-010-9329-x
Díaz-Leal JL, Torralbo F, Quiles FA, Pineda P, Alamillo JM (2014) Molecular and functional characterization of allantoate amidohydrolase from Phaseolus vulgaris. Physiol Plant 152:43–58. https://doi.org/10.1111/ppl.12157
Die JV, Román B, Nadal S, González-Verdejo CI (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232:145–153. https://doi.org/10.1007/s00425-010-1158-1
Dresler S, Hawrylak-Nowak B, Kováčik J, Pochwatka M, Hanaka A, Strzemski M, Sowa I, Wójciak-Kosior M (2019) Allantoin attenuates cadmium-induced toxicity in cucumber plants. Ecotoxicol Environ Saf 170:120–126. https://doi.org/10.1016/j.ecoenv.2018.11.119
Duran VA, Todd CD (2012) Four allantoinase genes are expressed in nitrogen-fixing soybean. Plant Physiol Biochem 54:149–155. https://doi.org/10.1016/j.plaphy.2012.03.002
Elstner EF, Heupel A (1976) Inhibition of nitrite formation from hydroxylammonium chloride: a simple assay for superoxide dismutase. Anal Biochem 70:616–620. https://doi.org/10.1016/0022-1759(78)90088-1
Forrester SJ, Kikuchi DS, Hernandes MS, Xu Q, Griendling KK (2018) Reactive oxygen species in metabolic and inflammatory signaling. Circ Res 122:877–902. https://doi.org/10.1161/circresaha.117.311401
Gus’kov EP, Kletskii ME, Kornienko IV, Olekhnovich LO, Chistyakov VA, Shkura TP, Prokof’ev VN, Zhdanov YA (2002) Allantoin as a free-radical scavenger. Dokl Biochem Biophys 383:105–107. https://doi.org/10.1023/a:1015331601169
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplast I. kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. https://doi.org/10.1016/j.abb.2022.109248
Hesberg C, Hänsch R, Mendel RR, Bittner F (2004) Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J Biol Chem 279:13547–13554. https://doi.org/10.1074/jbc.M312929200
Hoagland DR, Arnon DI (1938) The water culture method for growing plants without soil. Univ Calif Agric Exp Stn Circ 347:1–39
Irani S, Todd CD (2016) Ureide metabolism under abiotic stress in Arabidopsis thaliana. J Plant Physiol 199:87–95. https://doi.org/10.1016/j.jplph.2016.05.011
Irani S, Lobo JM, Gray GR, Todd CD (2018) Allantoin accumulation in response to increased growth irradiance in Arabidopsis thaliana. Biol Plant 62:181–187. https://doi.org/10.1007/s10535-017-0747-2
Jakše B, Jakše B, Pajek M, Pajek J (2019) Uric acid and plant-based nutrition. Nutrients 11:1736. https://doi.org/10.3390/nu11081736
Jana S, Choudhuri MA (1981) Glycolate metabolism of three submerged aquatic angiosperms during aging. Aquat Bot 12:345–354. https://doi.org/10.1016/0304-3770(82)90026-2
Janků M, Luhová L, Petřivalský M (2019) On the origin and fate of reactive oxygen species in plant cell compartments. Antioxidants 8:105. https://doi.org/10.3390/antiox8040105
Jung HI, Lee BR, Chae MJ, Lee EJ, Lee TG, Jung GB, Kim MS, Lee J (2020) Ascorbate-mediated modulation of cadmium stress responses: reactive oxygen species and redox status in Brassica napus. Front Plant Sci 11:586547. https://doi.org/10.3389/fpls.2020.586547
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lambert R, Quiles FA, Galvez-Valdivieso G, Piedras P (2017) Nucleases activities during French bean leaf aging and dark-induced senescence. J Plant Physiol 218:235–242. https://doi.org/10.1016/j.jplph.2017.08.013
Lamberto I, Percudani R, Gatti R, Folli C, Petrucco S (2010) Conserved alternative splicing of Arabidopsis transthyretin-like determines protein localization and S-allantoin synthesis in peroxisomes. Plant Cell 22:1564–1574. https://doi.org/10.1105/tpc.109.070102
Lescano CI, Martini C, González CA, Desimone M (2016) Allantoin accumulation mediated by allantoinase downregulation and transport by ureide permease 5 confers salt stress tolerance to Arabidopsis plants. Plant Mol Biol 91:581–595. https://doi.org/10.1007/s11103-016-0490-7
Lescano I, Devegili AM, Martini C, Tessi TM, González CA, Desimone M (2020) Ureide metabolism in Arabidopsis thaliana is modulated by C:N balance. J Plant Res 133:739–749. https://doi.org/10.1007/s10265-020-01215-x
Liu C, Guo J, Cui Y, Lü T, Zhang X, Shi G (2011) Effects of cadmium and salicylic acid on growth, spectral reflectance and photosynthesis of castor bean seedlings. Plant Soil 344:131–141. https://doi.org/10.1007/s11104-011-0733-y
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Ma X, Wang W, Bittner F, Schmidt N, Berkey R, Zhang L, King H, Zhang Y, Feng J, Wen Y, Tan L, Yue Li Y, Zhang Q, Deng Z, Xiong X, Xiao S (2016) Dual and opposing roles of xanthine dehydrogenase in defense-associated reactive oxygen species metabolism in Arabidopsis. Plant Cell 28:1108–1126. https://doi.org/10.1105/tpc.15.00880
Molins-Legua C, Meseguer-Lloret S, Moliner-Martinez Y, Campíns-Falcó P (2006) A guide for selecting the most appropriate method for ammonium determination in water analysis. TrAC Trends Anal Chem 25:282–290. https://doi.org/10.1016/j.trac.2005.12.002
Montalbini P (2000) Xanthine dehydrogenase from leaves of leguminous plants: purification, characterization and properties of the enzyme. J Plant Physiol 156:3–16. https://doi.org/10.1016/S0176-1617(00)80266-7
Nazar R, Iqbal N, Masood A, Khan MIR, Syeed S, Khan NA (2012) Cadmium toxicity in plants and role of mineral nutrients in its alleviation. Am J Plant Sci 3:1476–1489. https://doi.org/10.4236/ajps.2012.310178
Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138:304–318. https://doi.org/10.1104/pp.104.053793
Nourimand M, Todd CD (2016) Allantoin increases cadmium tolerance in Arabidopsis via activation of antioxidant mechanisms. Plant Cell Physiol 57:485–2496. https://doi.org/10.1093/pcp/pcw162
Pessoa J, Sárkány Z, Ferreira-da-Silva F, Martins S, Almeida MR, Li J, Damas AM (2010) Functional characterization of Arabidopsis thaliana transthyretin-like protein. BMC Plant Biol 10:30. https://doi.org/10.1186/1471-2229-10-30
Quiles FA, Raso MJ, Pineda M, Piedras P (2009) Ureide metabolism during seedling development in French bean (Phaseolus vulgaris). Physiol Plantarum 135:19–28. https://doi.org/10.1111/j.1399-3054.2008.01173.x
Quiles FA, Galvez-Valdivieso G, Guerrero-Casado J, Pineda M, Piedras P (2019) Relationship between ureidic/amidic metabolism and antioxidant enzymatic activities in legume seedlings. Plant Physiol Biochem 138:1–8. https://doi.org/10.1016/j.plaphy.2019.02.016
Raso MJ, Pineda M, Piedras P (2007) Tissue abundance and characterization of two purified proteins with allantoinase activity from French bean (Phaseolus vulgaris). Physiol Plant 131:355–366. https://doi.org/10.1111/j.1399-3054.2007.00969.x
Rothe GM (1974) Aldehyde oxidase isoenzymes (EC 1.2.3.1) in potato tubers (Solanum tuberosum). Plant Cell Physiol 15:493–499. https://doi.org/10.1093/oxfordjournals.pcp.a075029
Sagi M, Omarov RT, Lips SH (1998) The Mo-hydroxylases xanthine dehydrogenase and aldehyde oxidase in ryegrass as affected by nitrogen and salinity. Plant Sci 135:125–135. https://doi.org/10.1016/S0168-9452(98)00075-2
Schubert KR (1986) Products of biological nitrogen fixation in higher plants: synthesis, transport, and metabolism. Annu Rev Plant Biol 37:539–574. https://doi.org/10.1146/annurev.pp.37.060186.002543
Serventi F, Ramazzina I, Lamberto I, Puggioni V, Gatti R, Percudani R (2010) Chemical basis of nitrogen recovery through the ureide pathway: formation and hydrolysis of S-ureidoglycine in plants and bacteria. ACS Chem Biol 5:203–214. https://doi.org/10.1021/cb900248n
Soltabayeva A, Srivastava S, Kurmanbayeva A, Bekturova A, Fluhr R, Sagi M (2018) Early senescence in older leaves of low nitrate-grown atxdh1 uncovers a role for purine catabolism in N supply. Plant Physiol 178:1027–1044. https://doi.org/10.1104/pp.18.00795
Soltabayeva A, Bekturova A, Kurmanbayeva A, Oshanova D, Nurbekova Z, Srivastava S, Standing D, Sagi M (2022) Ureides are similarly accumulated in response to UV-C irradiation and wound but differently remobilized during recovery in Arabidopsis leaves. J Exp Bot 73:1016–1032. https://doi.org/10.1093/jxb/erab441
Stroiński A, Giżewska K, Zielezińska M (2013) Abscisic acid is required in transduction of cadmium signal to potato roots. Biol Plant 57:121–127. https://doi.org/10.1007/s10535-012-0135-x
Sun T, Pei T, Yang L, Zhang Z, Li M, Liu Y, Ma F, Liu C (2021) Exogenous application of xanthine and uric acid and nucleobase-ascorbate transporter MdNAT7 expression regulate salinity tolerance in apple. BMC Plant Biol 21:52. https://doi.org/10.1186/s12870-021-02831-y
Takagi H, Watanabe S, Tanaka S, Matsuura T, Mori IC, Hirayama T, Shimada H, Sakamoto A (2018) Disruption of ureide degradation affects plant growth and development during and after transition from vegetative to reproductive stages. BMC Plant Biol 18:287. https://doi.org/10.1186/s12870-018-1491-2
Todd CD, Polacco JC (2006) AtAAH encodes a protein with allantoate amidohydrolase activity from Arabidopsis thaliana. Planta 223:1108–1113. https://doi.org/10.1007/s00425-006-0236-x
Todd CD, Tipton PA, Blevins DG, Piedras P, Pineda M, Polacco JC (2006) Update on ureide degradation in legumes. J Exp Bot 57:5–12. https://doi.org/10.1093/jxb/erj013
Vogels GD, Van der Drift C (1970) Differential analysis of glyoxylate derivatives. Anal Biochem 33:143–157. https://doi.org/10.1016/0003-2697(70)90448-3
Wang P, Kong CH, Sun B, Xu XH (2012) Distribution and function of allantoin (5-ureidohydantoin) in rice grains. J Agric Food Chem 60:2793–2798. https://doi.org/10.1021/jf2051043
Watanabe S, Nakagawa A, Izumi S, Shimada H, Sakamoto A (2010) RNA interference-mediated suppression of xanthine dehydrogenase reveals the role of purine metabolism in drought tolerance in Arabidopsis. FEBS Lett 584:1181–1186. https://doi.org/10.1016/j.febslet.2010.02.023
Watanabe S, Matsumoto M, Hakomori Y, Takagi H, Shimada H, Sakamoto A (2014) The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ 37:1022–1036. https://doi.org/10.1111/pce.12218
Wellburn RW (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144:307–313. https://doi.org/10.1016/S0176-1617(11)81192-2
Wells XE, Lees EM (1991) Ureidoglycolate amidohydrolase from developing French bean fruits (Phaseolus vulgaris L.). Arch Biochem Biophys 287:151–159. https://doi.org/10.1016/0003-9861(91)90400-d
Werner AK, Witte CP (2011) The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci 16:381–387. https://doi.org/10.1016/j.tplants.2011.03.012
Werner AK, Sparkes IA, Romeis T, Witte CP (2008) Identification, biochemical characterization, and subcellular localization of allantoate amidohydrolases from Arabidopsis and soybean. Plant Physiol 146:418–430. https://doi.org/10.1104/pp.107.110809
Werner AK, Medina-Escobar N, Zulawski M, Sparkes IA, Cao FQ, Witte CP (2013) The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase in Arabidopsis, soybean, and rice. Plant Physiol 163:672–681. https://doi.org/10.1104/pp.113.224261
Witte CP, Herde M (2020) Nucleotide metabolism in plants. Plant Physiol 182:63–78. https://doi.org/10.1104/pp.19.00955
Yi SY, Lee M, Rameneni JJ, Lu L, Kaur C, Lim YP (2021) Xanthine-derived metabolites enhance chlorophyll degradation in cotyledons and seedling growth during nitrogen deficient condition in Brassica rapa. Plant Signal Behav 16:1913309. https://doi.org/10.1080/15592324.2021.1913309
You S, Zhu B, Wang F, Han H, Sun M, Zhu H, Peng R, Yao Q (2017) A Vitis vinifera xanthine dehydrogenase gene, VvXDH, enhances salinity tolerance in transgenic Arabidopsis. Plant Biotechnol Rep 11:147–160. https://doi.org/10.1007/s11816-017-0437-8
Zdunek-Zastocka E, Lips HS (2003a) Is xanthine dehydrogenase involved in response of pea plants (Pisum sativum L.) to salinity or ammonium treatment? Acta Physiol Plant 25:395–401. https://doi.org/10.1007/s11738-003-0021-4
Zdunek-Zastocka E, Lips HS (2003b) Plant molybdoenzymes and their response to stress. Acta Physiol Plant 25:437–452. https://doi.org/10.1007/s11738-003-0026-z
Zdunek-Zastocka E, Grabowska A, Michniewska B, Orzechowski S (2021) Proline concentration and its metabolism are regulated in a leaf age dependent manner but not by abscisic acid in pea plants exposed to cadmium stress. Cells 10:946. https://doi.org/10.3390/cells10040946
Zhang H, Jiang C, Lei J, Dong J, Ren J, Shi X, Zhong C, Wang X, Zhao X, Yu H (2022) Comparative physiological and transcriptomic analyses reveal key regulatory networks and potential hub genes controlling peanut chilling tolerance. Genomics 114:110285. https://doi.org/10.1016/j.ygeno.2022.110285
Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57:805–836. https://doi.org/10.1146/annurev.arplant.57.032905.105421
Acknowledgements
The authors would like to thank Magdalena Paradowska for technical assistance in the determination of ureides.
Funding
This research was partially founded by WULS-SGGW grant number S00125/2020.
Author information
Authors and Affiliations
Contributions
EZZ conceived and designed the experiments; EZZ, AG, and BM performed the analyses; EZZ, SO, JC, JF, MS analyzed the data, EZZ wrote the manuscript; JF and MS revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Fangjie Zhao.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 995 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zdunek-Zastocka, E., Grabowska, A., Michniewska, B. et al. Phaseolus vulgaris and Pisum sativum, representing ureide and amide legumes respectively, exploit ureides differentially to mitigate the deleterious effects of cadmium toxicity. Plant Soil 492, 439–456 (2023). https://doi.org/10.1007/s11104-023-06188-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06188-z