Abstract
Background and aims
Meloidogyne arenaria is an economically important root-knot nematode species. Successful plant infection by nematode is facilitated by parasite effectors. This study aimed to characterize a candidate M. arenaria effector, indicate its molecular partners from maize, and analyze its role during infection.
Material and methods
At first, we performed EST database mining to find candidate effector protein from M. arenaria. The expression of its coding gene in nematode developmental stages was assessed using digital droplet PCR. Candidate effector molecular partners were determined using yeast two-hybrid screening of maize cDNA library and interactions were confirmed by co-immunoprecipitation after co-expression in Nicotiana benthamiana. Candidate effector and its molecular partners were GFP-fused and localization in N. benthamiana leaves was observed under confocal microscope. Then, expression level of genes encoding interacting proteins from maize was measured.
Results
MaMsp4 protein was evaluated as candidate effector in M. arenaria and the highest expression level of its coding gene was observed in stage J2. MaMsp4 maize molecular partners were indicated, interactions with beta-galactosidase 11, pectinesterase, S-adenosyl methionine decarboxylase 2, and ethanolamine-phosphate cytidylyltransferase were confirmed, and all proteins fused with GFP were detected in the apoplast and/or cytoplasm. Genes of beta-galactosidase 11 and pectinesterase, playing role in cell wall modifications, were overexpressed at 24 hpi followed by down-regulation at 7 dpi, while S-adenosyl methionine decarboxylase 2 and ethanolamine-phosphate cytidylyltransferase, involved in plant defense response, were suppressed at 7 dpi, without preceding up-regulation.
Conclusions
We have found that MaMsp4 interacts with plant proteins involved in plant cell wall modifications and defense mechanisms related to polyamines biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meloidogyne arenaria, next to M. hapla, M. incognita, and M. javanica, is one of the most invasive root-knot nematode (RKN) species (Jones and Goto 2011). This highly polyphagous nematode infects both, mono- and dicotyledonous plants, including many crop species, with maize (Zea mays L.) as one of its main monocotyledonous hosts (CABI 2022; Przybylska and Obrępalska-Stęplowska 2020). Symptoms of RKN infection are manifested mostly in the underground parts with galls on roots and tubers (CABI 2022). During the RKN’s invasion, transcriptional reprogramming occurs, resulting in giant cells formation, which is necessary for these nematodes to feed, evolve and reproduce (Jones 1981).
During the host response to Meloidogyne infection, the expression of genes encoding proteins involved in many metabolic pathways is induced (Gheysen and Fenoll 2002; Przybylska et al. 2018). The first layers of the plant defense response constitute physical barriers as e.g., cell walls, and when they are overcome, other protection mechanisms are activated (Malinovsky et al. 2014). Next, when the plant cell pattern recognition receptors (PRRs) identify pathogen-associated molecular patterns (PAMPs) from the nematode, PAMPs-triggered immunity (PTI) is induced. Many PRRs have been described so far for numerous plant pathogens (Ngou et al. 2022). PTI may be suppressed by pest effectors, which consequently activates the effector-triggered susceptibility (ETS). On the other hand, when products of plant resistance genes interact with proteins secreted by the nematode, effector-triggered immunity (ETI) is induced. When the plant host develops disease resistance, ETI is accelerated, and the PTI is activated again (Jones and Dangl 2006). In these interactions, the nematode effectors play a key role.
The mentioned nematode effectors are small proteins, secreted in the nematode gland cells and injected by larvae in the J2 invasive stage through stylet into the plant tissue, resulting in the alterations of host-cell structure and function (Mitchum et al. 2013). Nematode effectors may be secreted by subventral gland cells (SvG) and by the dorsal esophageal gland cells (DG) (Mejias et al. 2019). The effectors, allowing J2 penetration and migration, are secreted in SvG, while the proteins, secreted during parasitism, are produced by both, SvG and DG (Nguyen et al. 2018). Moreover, as a result of recent studies, candidate effector proteins coding genes in adult females’ DG that may play role in later stages of parasitism were also indicated (Rocha et al. 2023).
Many RKN effectors and their functions taking part in the interaction with different host species have been described so far (Przybylska and Obrępalska-Stęplowska 2020; Vieira and Gleason 2019). The effectors secreted by RKNs are classified into groups, depending on their function during infection: plant cell wall degrading enzymes, plant defense modulators, plant hormone regulators, cell cycle modulators, cytoskeleton organizers, and plant metabolic re-programmers (Jagdale et al. 2021). They are involved, among others, in suppressing PTI response, like in the case of the Mh265 protein in M. hapla (Gleason et al. 2017), Mg01965 and MgMO237 in M. graminicola (Chen et al. 2018; Zhuo et al. 2019), or MiMIF-2 in M. incognita (Zhao et al. 2020). Furthermore, they can be assigned specifically to the salicylic acid (SA) pathway, as MiCM3, Minc03329, and MiPDCD6 effectors (Kamaruzzaman et al. 2023; Wang et al. 2018; Zhou et al. 2023) or to the jasmonic acid (JA) pathway as MiSE6 (Shi et al. 2018a), all from M. incognita. Moreover, the effectors can suppress ETI responses, as M. enterolobii protein – MeTCTP (Guo et al. 2022; Zhuo et al. 2017), M. graminicola - MgGPP (Chen et al. 2017), and M. javanica - MjShKT (Kumar et al. 2023) or suppress both, PTI and ETI responses as Msp40 from M. incognita (Niu et al. 2016) or Msp18, effector, common for different Meloidogyne species (Grossi-de-Sa et al. 2019).
On the other hand, some of the RKN effectors are involved in reactive oxygen species (ROS) burst, e.g. MgMO289 and MgPDI M. graminicola proteins (Song et al. 2021; Tian et al. 2019) as well as MiGST_N_4, MiEFh, MiACPS and MiTSPc from M. incognita (Pu et al. 2022), and MjNEROSs from M. javanica (Stojilkovic et al. 2022). Some others have completely different functions, e.g., the MiPFN3 protein from M. incognita binds plant actin monomers to manipulate this protein’s function (Leelarasamee et al. 2018). Moreover, downregulation of Minc03328 effector gene from M. incognita, triggered in A. thaliana by RNAi silencing, strongly reduced plant susceptibility to this nematode (Moreira et al. 2022). There are also effectors with multiple roles in the different processes as the Mj-FAR-1 protein, secreted by M. javanica in sedentary stages (Iberkleid et al. 2015). In tomato line overexpressing the gene encoding Mj-FAR-1 protein together with JA-responsive genes most of the cell wall modification-related genes were downregulated, which indicates the Mj-FAR-1 role in suppression of plant immune response. Another known effector playing a role in suppressing basal plant immunity is MgMo237 protein from M. graminicola. This effector interacts with three plant proteins involved in plant defense, consequently leading to the suppression of plant-defense genes, cell wall callose deposition, and ROS burst (Chen et al. 2018).
Nematode effectors may localize in different cell compartments such as the nucleus (Lin et al. 2013), cytoplasm (Gleason et al. 2017), apoplast (Tian et al. 2019), or even plastids (Lin et al. 2016). The effector may interact with more than one protein in the plant, which was also described for other plant pathogens such as bacteria, in infection with which the effectors may have a few targets with similar or different functions (Khan et al. 2018). The effector function may differ depending on the type of molecular partners it interacts with at different infection stages (Thordal-Christensen et al. 2018).
For M. arenaria, only an MA-CM-1 was suggested to act as an effector protein, on the basis of its sequence similarity to the other described nematode effectors, but its cellular localization and function have not been determined yet (Long et al. 2006). Moreover, there are no data describing the role of any RKN effector involved in RKN’s interactions with maize – one of the staple crops produced worldwide. Therefore, this study was aimed at identification of a potential M. arenaria effector protein involved in the maize – M. arenaria interactions, determination of its expression pattern during developmental stages, localization in the plant cell, as well as indication of its molecular partners from the host plant to demonstrate its function during parasitism.
Material and methods
Plant material, nematode samples, and bacteria strains
The nematode material used in this study was M. arenaria population, derived from the Institute of Plant Protection – NRI collection, harvested in four different life stages: eggs, J2 invasive larvae, J3/J4 invasive larvae, and adult females. Moreover, one variety of maize (PR39F58, Pioneer) was used, with high susceptibility to RKN infection, evaluated previously (Przybylska et al. 2018). The nematode eggs were extracted from maize roots using the technique described by Hussey (1973), J2 larvae were hatched from eggs and J3/J4 larvae, and adult females were extracted from infected maize plants at 3 and 6 weeks after infection, respectively, using the method described by Mani et al. (2020). In the J3/J4 larvae mixture, the ratio between J3 and J4 was not determined because of the high similarity between these two stages. All experiments with maize plants were maintained under constant greenhouse conditions, with day/night temperatures 26 °C /21 °C, air humidity 50%, and additional lighting from 7 am to 7 pm. To carry out the localization and co-immunoprecipitation (Co-IP) assays, Nicotiana benthamiana plants and two Agrobacterium tumefaciens strains: C58C1 and EHA105 were used. A. tumefaciens strains were selected to provide the highest efficiency for the expression vectors used in these assays. For the pAUL1 vector, the highest efficiency was observed with the C58C1 strain and for the pBin61 vector with the EHA105 strain (unpublished data).
Search for a potential M. arenaria effector through data mining and its in-silico analysis
To determine the potential M. arenaria effector, comparative BLAST analyses were carried out between a database prepared from the sequences encoding M. incognita potential effectors, analyzed by Huang et al. (2003) and EST sequences database from M. arenaria, deposited in the National Center for Biotechnology Information (NCBI) database. The selected sequence was also used to query the WormBase Parasite database with the BLASTN algorithm (at www.parasite.wormbase.org).
The gene encoding the Msp4 effector protein from M. incognita (AF531163) was selected for further analyses. Because of its high similarity to M. arenaria EST (94.81%), the Msp4 coding sequence was used as a template for the primers’ design. To amplify the entire Msp4 coding sequence in the M. arenaria species, forward (Msp4F: GCAACTTTTATTGCGTA) and reverse (Msp4R: GTTTTACTCATCAGCACA) primers were designed upstream and downstream the ATG and stop codons, respectively. PCR was performed using the cDNA synthesized from RNA, extracted from M. arenaria in the J2 invasive stadium. Total RNA was extracted from juveniles, using Total RNA Mini Concentrator (A&A Biotechnology, Gdańsk, Poland). One-hundred nanograms of RNA were used as a template for cDNA synthesis with Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific, Waltham, USA) in a final volume of 20 μl.
RT-PCR was carried out in a reaction mixture containing: 5 μl 2x DreamTaq MasterMix (Thermo Fisher Scientific, Waltham, USA), 0.1 μM of each primer, 1 μl of cDNA template, and water up to 10 μl. A no-template control reaction was performed as well. The reaction was run with the following thermal profile: 5 min at 95 °C; 30 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, and final extension for 5 min at 72 °C. PCR products were separated in 1% agarose gel with Simply Safe stain (EURx, Gdańsk, Poland) and visualized under UV light. The PCR product of expected size was then excised from the gel, eluted using Wizard® SV Gel and PCR Clean-Up System (Promega, Walldorf, Germany), and ligated with pJET 1.2 vector (Thermo Fisher Scientific, Waltham, USA). The recombined plasmid was transformed into Escherichia coli DH10B strain, amplified, isolated from bacteria cells using NucleoSpin Plasmid Kit (Macherey-Nagel, Düren, Germany), sequenced, and finally compared with the other sequences encoding Meloidogyne spp. effectors and analyzed in BioEdit software (Hall 1999).
Protein structure model was predicted using ColabFold v1.5.2 software (Mirdita et al. 2022). The ColabFold uses AlphaFold2 (Bryant et al. 2022), an innovative tool for protein modelling, while datasets analysis and sequence alignments are generated through MMseqs2 (Steinegger and Söding 2017).
Digital droplet PCR analysis of MaMsp4 gene expression level
The number of copies of MaMsp4-derived transcripts in the samples derived from M. arenaria in 4 different life stages: eggs, larvae J2, larvae J3/J4 (mixture of both stages) and adult females. Was evaluated using digital droplet PCR (ddPCR). Each stage was analyzed in three biological replicates and 100 ng of RNA from each sample, extracted from specimens using Total RNA Mini Concentrator (A&A Biotechnology, Gdańsk, Poland), were taken for cDNA synthesis with a Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific, Waltham, USA) in 20 μl final volume. Primers, amplifying an internal fragment of MaMsp4 gene: MaMsp4RTF (ACCGTGGAAAGCGTCTTAAC) and MaMsp4RTR (TCAATTTTGCCGCGCATCTTT) were designed.
All reactions were run in 20 μl reaction volume containing 10 μl of 2x QX200 EvaGreen Digital PCR Supermix (Bio-Rad, Hercules, USA), 0.1 μM of each primer, 1 μl of cDNA template, and water, as described previously (Budziszewska et al. 2021). A sample without the cDNA template was used as a negative control. Each sample was mixed with 20 μl of Droplet Generation Oil for Probes (Bio-Rad, Hercules, USA) and droplets were created using QX200 Droplet Generator (Bio-Rad, Hercules, USA). The PCR reaction was performed with the following thermal profile: 5 min at 95 °C; 38 cycles of 30 s at 95 °C and 1 min at 60 °C, cooling to 4 °C for 5 min, and termination of the reaction for 5 min at 90 °C. The fluorescence signal was assessed with QX200 AutoDG System (Bio-Rad, Hercules, USA), to distinguish positive and negative droplets.
Yeast two-hybrid screening of maize cDNA library
The yeast two-hybrid (Y2H) screening was performed by Creative BioLabs Inc. (Shirley, USA) using Matchmaker Gold System (Takara Bio Inc., Kusatsu, Japan). The bioinformatical analysis of the full-length MaMsp4 protein, using the SignalP tool (Petersen et al. 2011), was carried out and indicated that it is a secreted protein with a signal peptide. The first 24 amino acids of the N-terminus of MaMsp4 protein, containing a hydrophobic structure, were excluded, because the full-length protein with the signal peptide may be difficult to keep in cells, as described by Creasey et al. (2003), and the truncated MaMsp4 sequence (named MaMsp4–24) was used in further analysis. The MaMsp4–24 sequence was subcloned into the bait vector (pGBKT7, Takara Bio Inc., Kusatsu, Japan), transformed into the yeast strain Y2HGold (Takara Bio Inc., Kusatsu, Japan) which was examined for toxicity and self-activation effects caused by the bait plasmid.
The maize cDNA library, constructed by Creative BioLabs Inc. (Shirley, USA) was customized using the pGADT7 vector (Takara Bio Inc., Kusatsu, Japan), and its quality was evaluated by transforming it to E. coli cells, amplification of 24 randomly selected clones, and amplicon sizes analyses. Y2H screening was performed by the mating method and the number of screened transformants was approximately of 2.5 × 107 colony-forming units. The potential positive hybrids were tested for histidine (His) and adenine (Ade) activation and selected positive hybrids were plated on a medium with X-a-Gal. For further confirmation of protein interactions, each potentially interacting prey plasmid was extracted, sequenced and those with unique protein-encoding genes were selected and co-transformed with the bait plasmid back into the yeast and tested again on a medium without His and Ade and with X-a-Gal.
Co-immunoprecipitation (co-IP) assay
For a Co-IP assay, the nematode candidate effector protein was fused with 3xHA tag, whereas the GFP was fused to the N-end of maize proteins. The sequence encoding MaMsp4 protein, without a stop codon, was amplified by RT-PCR, using primers listed in the table (Online Resource 1), cloned into the Gateway donor vector pDONRZeo using BP clonase (Thermo Fisher Scientific, Waltham, USA), and subcloned using Gateway LR clonase (Thermo Fisher Scientific, Waltham, USA) into the pAUL1 vector, having a C-terminal 3xHA tag coding sequence (Lyska et al. 2013). RT-PCR was used to amplify the coding sequence of these six MaMsp4 molecular partners, from which full coding sequences were available. Maize cDNA was synthesized with Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific, Waltham, USA) on the template of 100 ng of RNA and extracted using a Total RNA Mini Concentrator (A&A Biotechnology, Gdańsk, Poland). The sequences encoding these proteins were cloned into pJET1.2 vector (Thermo Fisher Scientific, Waltham, USA), transformed into E. coli cells, and after isolation with NucleoSpin Plasmid Kit (Macherey-Nagel, Düren, Germany) were subjected to subsequent Sanger sequencing. The coding sequences of the analyzed maize proteins were fused with the C-terminus of GFP and subcloned under control of 35S promoter and nos terminator in the pBin61 expressing vector (Bendahmane et al. 2002) using NEBuilder HiFi DNA Assembly Kit (New England Biolabs, Ipswich, USA). Primers used for this approach are listed in the table (Online Resource 1).
Prepared constructs were cloned in E. coli cells, isolated using a NucleoSpin Plasmid kit (Mecherey-Nagel, Düren, Germany), sequenced, and transformed into A. tumefaciens C58C1 strain (for pAUL1-derived plasmids) or EHA105 strain (for pBin61-derived constructs). A suspension of A. tumefaciens carrying the analyzed plasmids was grown at 28 °C for 24 h in an LB medium with kanamycin and rifampicin (EHA105 strain) or kanamycin and tetracycline (C58C1 strain), harvested, suspended in infiltration buffer (10 mM MES, pH 5.8, 0.5 μM acetosyringone, and 10 mM MgCl2) and kept in the dark for 4 h. Bacterial density was measured at OD600, adjusted to 1.0, and the suspension was used to infiltrate leaves of 4–6-week-old N. benthamiana plants. The leaves were infiltrated with a mixture of A. tumefaciens suspension carrying plasmids with the pairs of MaMsp4 protein with each of the analyzed plant proteins, mixed in the volume ratio of 1:1. Controls with a MaMsp4 and an empty vector, maize proteins with an empty vector as well as two empty vectors were also analyzed.
The agroinfiltrated leaves were collected 3 days after infiltration, pulverized in liquid nitrogen and 500 mg of plant material was taken for extraction, performed in 750 μl of buffer containing: 100 mM Tris-Cl pH 8,0; 2 mM EDTA; 2 mM EGTA; 150 mM NaCl; 1% CHAPS; 2 mM DTT and protease inhibitor cocktail (BioShop Canada Inc., Burlington, Canada). The homogenate was incubated with shaking at 4 °C for 30 min and centrifuged at 4 °C with 15,000 x g for 15 min. The supernatant was collected, and 50 μl of a Monoclonal Anti-HA-Agarose suspension (Thermo Fisher Scientific, Waltham, USA) was added to each sample and incubated for 1 h at 4 °C with gentle agitation. The slurry was centrifuged for 15 s at 12,000 x g, the supernatant was removed, the resin was washed four times with 1 ml of PBS buffer each time, and after the last wash, about 10–20 μl of PBS buffer was left above the beads. The beads were incubated with 30 μl of 2x Laemmli buffer (0.125 M Tris-Cl pH 6.8; 4% SDS; 20% glycerol; 10% 2-mercaptoethanol; 0.05% bromophenol blue) for 3 min at 95 °C, centrifuged for 15 s with 12,000 x g and the supernatant was collected to fresh tubes. Twenty microliters of each control sample were mixed with 20 μl of 2x Laemmli buffer and boiled for 5 min at 95 °C.
Leaves samples were analyzed using Western Blot with antibodies against HA and GFP. The samples were separated in the polyacrylamide (PAA) gel prepared using TGX™ FastCast™ Acrylamide Kit, 12% (Bio-Rad, Hercules, USA), and run in the electrophoresis buffer (0.1 M Tris; 0.4 M glycine; 0.1% SDS). Proteins from the gel were blotted to a PVDF membrane by the semi-dry transfer in the presence of Towbin buffer (25 mM Tris; 150 mM glycine; 10% methanol) using a Trans-BloT® Turbo™ Transfer System (Bio-Rad, Hercules, USA). The membranes were blocked overnight in a 5% skim milk solution in TBS-T buffer (10 mM Tris-Cl pH 7.5; 150 mM NaCl; 0.1% Tween-20) at 4 °C with shaking. Next, the filter was incubated for 1.5 h at room temperature in a solution of primary antibody in a concentration of 1:2000 for anti-GFP (ProteinTech Group Inc., Rosemont, USA) or anti-HA (Thermo Fisher Scientific, Waltham, USA), in 5% skim milk solution in TBS-T buffer, and washed four times in TBS-T buffer. Subsequently, the membranes were incubated for 1 h at room temperature in a 1:40,000 solution of secondary antibody conjugated with alkaline phosphatase (anti-mouse, Sigma-Aldrich, Saint Louis, USA) in a 5% skim milk solution in TBS-T buffer and washed four times in TBS-T buffer. Then, the membranes were incubated in the Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega, Madison, USA) until the expected signal developed. To confirm the same protein loading, PAA gels after transfer were stained with Coomassie brilliant blue.
Cellular localization of GFP-fused proteins using confocal microscopy
The constructs fused with GFP in pBin61, described previously in the Co-IP assay, were used for localization analysis of potential interacting plant proteins. The sequence encoding MaMsp4 candidate effector protein with a stop codon was cloned in fusion with GFP at the N-terminus into the pBin61 expression vector using the technique described above and with primers listed in table (Online Resource 1). All plasmids were transformed into A. tumefaciens strain EHA105 and transient overexpression was performed in N. benthamiana leaves, using a technique described above for Co-IP assay. Fluorescence was observed 3 days after infiltration under a confocal microscope (Carl Zeiss, Stuttgart, Germany) in the Laboratory of Electron and Confocal Microscopy at Adam Mickiewicz University (Poznań, Poland).
Assessment of gene expression level of genes encoding maize proteins by means of quantitative real-time PCR
The expression level of genes encoding potential interacting proteins from maize was determined using a quantitative real-time PCR assay (RT-qPCR). Maize plants, highly susceptible to M. arenaria infection (PR39F58, Pioneer) were inoculated with ca. 1500 M. arenaria J2 larvae. Samples from leaves and roots of four healthy and four infected plants were collected at three time points: 24 hours post inoculation (24 hpi), 3 days (3 dpi), and 7 days (7 dpi) post inoculation. The RNA was extracted as described above for the nematodes, and cDNA was synthesized using 250 ng of the RNA using a Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific, Waltham, USA). Real-time PCR primers were designed based on the sequences of interest deposited in NCBI and listed below (Table 1). For normalization, two reference genes: leunig and fpgs, evaluated previously for maize (Manoli et al. 2012) were used.
All real-time PCR reactions were run in a 10 μl reaction mixture containing 5 μl of 2x iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, USA), 0.1 μM of each primer, 1 μl of cDNA template, and water. No-template control reactions were run. Reactions were conducted in LightCycler 96 platform (Roche, Basel, Switzerland) under the following thermal profile: 5 min at 95 °C; 40 cycles of 20 s at 95 °C, 20 s at a temperature proper for each primers pair (Table 1.), and 20 s at 72 °C. The melting curve was generated by heating the samples starting from 65 °C to 95 °C, with at a rate of 1 °C. Relative quantification, normalized against the leunig and fpgs reference genes, was carried out with GenEx 6.0 software (MultiD Analyses AB) using the 2-ΔΔCq method. Statistical significance was calculated with the t-student test and P < 0.05 or with the Mann-Whitney test for non-parametric results.
Protein-protein interaction networks analysis
Interaction networks of selected maize proteins were analyzed using the STRING (Szklarczyk et al. 2019) database (www.string-db.org).
Results
Search for a potential M. arenaria effector and MaMsp4 protein coding sequence analysis
BLAST analysis of M. incognita effectors and M. arenaria ESTs sequences was performed and as a result, the EST sequences from M. arenaria with high similarity to the sequences encoding four M. incognita effector proteins were found with a score of at least 140 (Online Resource 2). Sequence encoding effector protein Msp4 (AF531163), whose longest M. arenaria EST sequence was available (664 bp), was selected to further analyses. Sequences analysis of the RT-PCR with Msp4F/R primers amplified 996 bp fragment of MaMsp4 (Fig. 1A) revealed its 96.6% and 90.2% identity with the corresponding DNA sequence of Msp4 effector from M. incognita and ME-3C06 effector from M. enterolobii, respectively (Fig. 1B). The sequence was deposited in GenBank under OQ407870 accession number. BLAST analysis of the MaMsp4 sequence in the WormBase Parasite database showed 75 hits with a score higher than 100 (Online Resource 3) for M. arenaria, M. incognita, M. javanica, and M. floridensis species in datasets PRJEB8714, PRJNA340324, and PRJEB6016.
Analysis of MaMsp4 effector protein from M. areneria. A. RT-PCR amplification of MaMsp4 coding sequence. The 996 bp amplification product was indicated. NTC - no template control; B. Result of comparative sequences analysis between nucleotide sequences encoding MaMsp4 protein from M. arenaria, Msp4 from M. incognita and ME-3C06 from M. enterolobii; D. MaMsp4 protein model predicted with ColabFold tool (Mirdita et al. 2022); pIDDT - single chain predictions confidence level
MaMsp4 gene expression level is the highest in the J2 stage of the M. arenaria
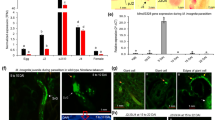
Effector genes are known to be highly expressed at the invasive J2 developmental stage. To assess the MaMsp4 gene expression in consecutive nematode developmental stages, ddPCR analysis was performed. The highest number of positive droplets was observed in samples from J2 larvae, followed by a similar number in samples from J3/J4 larvae and adult females, and the lowest number of transcripts was observed in the eggs’ samples. No positive droplets were obtained in the control sample without a cDNA template (Fig. 2A, B). An average number of transcripts was calculated for 1 μg of total RNA used in the reaction and the results were as follows: 34 copies/μg in eggs, 1754 copies/μg in J2 larvae, 250 copies/μg J3/J4 larvae, and 256 copies/μg in adult females (Fig. 2C).
Analysis of MaMsp4 gene expression across different M. arenaria developmental stages using droplet digital PCR (ddPCR) reaction. A. Graphical representation of positive and negative droplets; B. Number of positive droplets in each sample; C. Calculation of an average number of MaMsp4 transcripts in the samples from each developmental stage. NTC – no template control
Screening of maize cDNA library with yeast two-hybrid system with the MaMsp4 sequence as bait revealed its putative host molecular partners
Initial validation of the Zea mays cDNA library showed that the average length of the sequences was >800 bp and the positive rate of the library was >90%, which meets the requirements of a screening library to be used for subsequent Y2H assays (Fig. 3A). Neither toxicity nor self-activation effects were confirmed for any tested proteins. As a result of screening, on the plates without His and Ade, 24 positive clones were observed. All of these clones also were blue in the test with X-A-Gal (Fig. 3B). Sequence analysis showed that out of twenty-four positive clones, sixteen were carrying protein-coding genes (the remaining clones encoded non-protein sequences). All of these 16 positive clones were chosen for further confirmation of protein interactions (Table 2). Results of co-transformation of selected clones with the bait plasmid back into the yeast, showed that 7 clones, namely 2, 5, 10, 12, 14, 22, and 23, have reliable interactions with the bait protein (Fig. 3C). Accordingly, these 7 prey candidates were truly positive ones after validation.
Results of the maize cDNA library screening. A. Validation of the pre-made maize cDNA library; M: 2000 bp marker, lane 1–24: randomly selected clones. B. The final screening result of 24 positive clones. C. The speckle pattern of validation assay on medium with X-A-Gal added. Seven prey colonies (MaMsp4 + 2, 5, 10, 12, 14, 22, and 23) are blue, the same as the positive control. K+ − positive control, K- - negative control, MaMsp4 + EV – MaMsp4 + empty vector
Co-immunoprecipitation-based confirmation of interactions of the MaMsp4 with the selected molecular partners from maize
Co-expression of bait and putative prey proteins in N. benthamiana was followed by Co-IP assay to confirm their interactions in planta. In order to do this, 3 x HA-tagged MaMsp4 was co-expressed together with the GFP-tagged bait proteins, followed by incubation with anti-HA agarose. The eluted complexes were resolved in SDS-PAGE, transferred to PVDF and probed with anti-GFP antibodies. The expected 65 kDa band confirmed the production of 3 x HA-MaMsp4 in infiltrated leaves (Fig. 4A, lanes 1–7). On the other hand, in the same samples tested with anti-GFP antibody, the products were observed in lanes with samples containing MaMsp4 and its molecular partners from maize: beta-galactosidase 11 with size about 60 kDa (Fig. 4B, lane 1), S-adenosyl methionine decarboxylase 2 with size about 35 kDa (Fig. 4B, lane 2), pectinesterase with size about 70 kDa (Fig. 4B, lane 3), and ethanolamine-phosphate cytidylyltransferase with size about 80 kDa (Fig. 4B, lane 4). No specific product was observed in the samples containing MaMsp4 with polygalacturonase (Fig. 4B, lane 5) and uncharacterized protein LOC107325938 (Fig. 4B, lane 6). No band was also observed in the lanes with negative controls (Fig. 4B, lanes 7–13).
Results of western blot with samples after co-immunoprecipitation assay with antibody: A. anti-HA and B. anti-GFP. C. PAA gel stained with Coomassie blue as a loading control. 1 – beta-galactosidase 11 x MaMsp4; 2 – S-adenosyl methionine decarboxylase 2 x MaMsp4; 3 – pectinesterase x MaMsp4; 4 – ethanolamine-phosphate cytidylyltransferase x MaMsp4; 5 – polygalacturonase x MaMsp4; 6 – uncharacterized protein LOC107325938 x MaMsp4; 7 – pBin61empty x MaMsp4; 8 – beta-galactosidase 11 x pAUL1empty; 9 – S-adenosyl methionine decarboxylase 2 x pAUL1empty; 10 – pectinesterase x pAUL1empty; 11 – ethanolamine-phosphate cytidylyltransferase x pAUL1empty; 12 – polygalacturonase x pAUL1empty; 13 – uncharacterized protein LOC107325938 x pAUL1empty; 14 – pBin61empty x pAUL1empty
MaMsp4 protein and its molecular partners from maize localize at cytoplasm and apoplast
To indicate the cellular co-localization of MaMsp4 and its molecular partners, both the candidate effector and tested maize proteins were GFP-tagged and transiently expressed in N. benthamiana leaves. Three days after infiltration, 5 mm discs were cut from the leaves and scanned under a confocal microscope. Fluorescence was observed in the samples with vectors overexpressing GFP and GFP fused with all analyzed proteins, in contrast to the empty pBin61 vector (Fig. 5). For MaMsp4 protein, fluorescence was identified in the apoplast space and the cytoplasm (Fig. 5B). A similar observation was made for GFP-tagged S-adenosyl methionine decarboxylase 2 (Fig. 5D), ethanolamine-phosphate cytidylyltransferase (Fig. 5F), and uncharacterized protein LOC107325938 (Fig. 5H). Importantly, GFP-tagged beta-galactosidase 11 (Fig. 5C) resided mostly in the cytoplasm, whereas GFP-tagged pectinesterase (Fig. 5E) and polygalacturonase (Fig. 5G) were localized mainly in the apoplast.
Subcellular localization of MaMsp4 effector protein from Meloidogyne arenaria and its maize molecular partners, and potential interacting proteins (polygalacturonase and uncharacterized protein) fused with GFP at the N-terminus in pBin61 vector and overexpressed in N. benthamiana leaves. A. Positive control with plain GFP; B. MaMsp4; C. Beta-galactosidase 11; D. S-adenosyl methionine decarboxylase 2; E. Pectinesterase; F. Ethanolamine-phosphate cytidylyltransferase; G. Polygalacturonase; H. Uncharacterized protein LOC107325938; I. Empty pBin61 vector as a negative control. Bar = 100 μM
The expression level of genes encoding interacting proteins from maize is strongly suppressed at 7 dpi
Analysis of the expression of genes encoding maize proteins interacting with MaMsp4 candidate effector protein: beta-galactosidase 11, S-adenosyl methionine decarboxylase 2, pectinesterase, and ethanolamine-phosphate cytidylyltransferase, as well as potential interacting proteins – polygalacturonase and uncharacterized protein LOC107325938, revealed changes between the samples taken at different time points after nematode inoculation, as well as between the samples derived from leaves and roots (Fig. 6). The most significant differences were observed in the expression level of genes encoding polygalacturonase and beta-galactosidase 11, in which the statistically important down-regulation was observed at all time points in the leaf samples and 7 days after inoculation in the samples derived from roots. Moreover, down-regulation in the samples collected from roots at 7 dpi was found in all analyzed genes. On the other hand, statistically significant up-regulation in the root samples in a very early stage of infection (24 hpi) was observed in the genes encoding beta-galactosidase 11, pectinesterase, polygalacturonase as well as uncharacterized protein. Interestingly, in the leaf samples, downregulation was observed for all analyzed proteins at 24 hpi as well as at 7 dpi.
A. Relative expression levels of genes encoding maize proteins interacting, or potentially interacting with MaMsp4 effector protein. Asterisks indicate samples with statistically important up- or down-regulation (P < 0.05); B. Relative expression values with bolded statistically important results (P < 0.05)
Interaction networks analysis indicated that MaMsp4 interactions with S-adenosyl methionine decarboxylase 2 and ethanolamine-phosphate cytidylyltransferase might be functionally connected
Analysis of proteins interaction networks using the STRING database was carried out for MaMsp4 molecular partners: beta-galactosidase 11, S-adenosyl methionine decarboxylase 2, pectinesterase, and ethanolamine-phosphate cytidylyltransferase. Most of the results indicated the interaction networks associated with protein functions in the pathways in which they participate (Online resource 4, Online resource 5). The main interactors for S-adenosyl methionine decarboxylase 2 are spermidine synthases, while for ethanolamine-phosphate cytidylyltransferase - choline/ethanolamine kinases. However, the analysis revealed that ethanolamine-phosphate cytidylyltransferase, a key enzyme in the phosphatidylethanolamine biosynthesis pathway, interacts with S-adenosyl methionine, which is a substrate for S-adenosyl methionine decarboxylase in polyamine biosynthesis pathway (Fig. 7).
Discussion
Effector proteins are a key RKN weapon during plant colonization. They are secreted through the esophageal glands and injected by stylet to manipulate host cell metabolism (Jagdale et al. 2021). Although there are many studies characterizing their function in parasitism, there are still a lot of nematode species and their hosts for which the role of effectors has not been analyzed yet. This is true for M. arenaria as well. Moreover, the effectors and their role during RKN interaction with the maize, have also not been described as yet.
In this study, our aim was to identify M. arenaria candidate effector protein and describe its involvement in compatible interactions with maize. To achieve this, we performed BLAST analysis of M. incognita effectors and M. arenaria ESTs sequences and indicated MaMsp4 as a potential effector protein on the basis of its high identity level with the other two Meloidogyne spp. effectors: Msp4 from M. incognita and ME-3C06 protein from M. enterolobii. It is known that the Msp4 effector protein is expressed in DG in the parasitic J2 stage in M. incognita (Huang et al. 2003), which indicates its involvement in parasitism. The ME-3C06 protein sequence from M. enterolobii is deposited in the NCBI database and described as an effector protein encoding sequence, but, unfortunately, no further data have been published as yet.
Next, we analyzed the expression pattern of the MaMsp4 coding gene in different M. arenaria developmental stages. The ddPCR assay showed that the MaMsp4 expression was seven times higher in the samples derived from the J2 stage compared to those from J3/J4 and adult stages and about fifty times higher compared to those from eggs. This indicates the important role of this nematode protein in the invasive stage of parasitism. The results obtained for the Mc16D10L effector protein gene from M. chitwoodi (Dinh et al. 2014), MgGPP from M. graminicola (Chen et al. 2017), or Mi-msp2 from M. incognita (Joshi et al. 2019) indicated that the highest expression level of the genes encoding the corresponding effector proteins were observed in the invasive J2 stage as well.
The next step of the study was to identify the intracellular localization of the MaMsp4 protein. By analyzing transient expression of GFP-tagged MaMsp4 in N. benthamiana, followed by confocal microscopy scanning we observed that the MaMsp4-GFP is localized at the cytoplasm and apoplast. An analogous cellular localization has been observed for Mg16820 protein, an effector from M. graminicola, whose function is determined by localization. Namely, when it is localized in the apoplast, it is involved in the suppression of the PTI response. However, the same protein is involved in ETI suppression when accumulated in the cytoplasm (Naalden et al. 2018). Noticeably, some effectors were found only in the apoplast. Indeed, the MiCRT (Jaouannet et al. 2013) or Mg01965 (Zhuo et al. 2019) of M. incognita and M. graminicola, respectively, were described with apoplastic localization and are involved in the suppression of PTI.
Protein-protein interactions participate in all biological processes that may differ depending on the composition of the formed complex. Therefore, in the next step of resolving the biological function of the MaMsp4, we aimed at identification of its molecular partners from maize by Y2H screening. As a result, we indicated six proteins: beta-galactosidase 11, S-adenosyl methionine decarboxylase 2, pectinesterase, ethanolamine-phosphate cytidylyltransferase, polygalacturonase, and one protein with uncharacterized function (LOC107325938). Among these records, in the subsequent Co-IP assay, the interactions were confirmed for four proteins, the beta-galactosidase 11, S-adenosyl methionine decarboxylase 2, pectinesterase, and ethanolamine-phosphate cytidylyltransferase. No results observed in western blot with anti-GFP antibody in samples expressing the remaining two proteins: polygalacturonase and uncharacterized protein (LOC107325938) did not exclude interactions, but might rather be a consequence of low amounts of these proteins in the plant tissue after overexpression or other factors such as extraction difficulties.
Next, we performed the cellular localization analysis also for the proteins that we identified in the previous step as MaMsp4 interactors. We observed that the tested proteins were mainly localized in the cytoplasm (beta-galactosidase 11), apoplast (pectinesterase, polygalacturonase), or both cytoplasm and apoplast (S-adenosyl methionine decarboxylase 2, ethanolamine-phosphate cytidylyltransferase, and uncharacterized protein). Similar subcellular localization in the MaMsp4 protein indicates the possibility of interaction.
The first group of proteins interacting or potentially interacting with MaMsp4 includes beta-galactosidase 11, pectinesterase, and polygalacturonase, which are known to play a role in cell wall modifications and are crucial for successful feeding site formation and RKN development. The RKNs are capable of modulating the expression of many cell wall-modifying enzymes in the plant, including expansins, endoglucanases, extensins, hydrolases, and many others (Bozbuga et al. 2018; Petitot et al. 2020; Sobczak et al. 2011; Veronico et al. 2022).
However, beta-galactosidases in plants are involved in the degradation of structural polysaccharides in plant cell walls are also implicated in other biological processes (Hossain 2022; Seddigh and Darabi 2014). In maize, they have been reported to play an important role in energy generation and to be significantly depressed under abiotic stress conditions (Gao et al. 2020). The role of plant beta-galactosidases in nematode development was investigated through the interactions between M. incognita and A. thaliana. The nematode specimen number and size were reduced in A. thaliana mutant lacking a cell-wall-localized beta-galactosidase (Bozbuga et al. 2018). We observed a significant up-regulation of this gene in the very early stage of infection (24 hpi). Moreover, a strong down-regulation of the gene encoding beta-galactosidase in wheat roots 8 days after Heterodera avenae infection has been reported (Qiao et al. 2019). It is in line with our results showing the down-regulation 7 days after infection in the samples derived both, from leaves and roots.
Moreover, another important enzyme involved in cell wall modification, a molecular partner for MaMsp4 protein, is pectinesterase which plays an important role in cell wall metabolism, through degradation of pectin, one of the main components of the plant cell wall (Passianotto et al. 2017). Moreover, during the response of A. thaliana and soybean to M. incognita infection, significant overexpression of the genes encoding pectinesterase was observed in all stages of infection (Ibrahim et al. 2011). The above-mentioned genes are among the most frequently up-regulated ones upon cell wall penetration by a nematode (Passianotto et al. 2017), which is in agreement with the results observed in this study in the samples derived from roots, in which significant up-regulation occurred in the very early stage of infection. On the other hand, the inhibition of plant pectinesterases was shown to increase plant resistance to pathogens, for example, M. incognita (Gorshkov and Tsers 2022; Pham et al. 2013).
On the other hand, the degradation of pectins in the cell wall is catalyzed by polygalacturonases (He et al. 2019), which also were analyzed in this study as potentially interacting with the MaMsp4 candidate effector protein. Polygalacturonases have been also reported to be associated with the control of cell growth and development, wound responses, and plant-pathogen interactions (He et al. 2023; Kim et al. 2006; Lorrai and Ferrari 2021). The study of soybean response to H. glycines indicated that the higher accumulation of the polygalacturonase transcripts level in roots in the early stage of nematode infection could facilitate successful parasitism (Mahalingam et al. 1999), which is consistent with the results of our previous research in which a significant up-regulation was observed in the early stage of maize infection by M. arenaria (Przybylska and Spychalski 2021), which was additionally confirmed by the results of this study. Moreover, the enhanced expression of the polygalacturonase coding gene in maize leads to suppression of the cell death, caused by SA and 10-oxo-11-phytoenoic acid. On the other hand, the transient overexpression of this gene in N. benthamiana induced hypersensitive response (He et al. 2019). Noteworthy, polygalacturonases and pectinesterases were overexpressed in giant cells in rice after M. graminicola infection (Ji et al. 2013), which, in conjunction with our results, suggests their important role in the very early stage of infection, when feeding sites are formed.
Many nematode effectors are suppressing the plant defense response (Goode and Mitchum 2022). RKNs are able to manipulate the plant immune system by suppressing the SA-related pathway as described for M. incognita Mi-CM-3 effector, whose overexpression caused a reduction in the levels of SA and mRNA of gene encoding pathogenesis-related protein 1 in N. benthamiana (Wang et al. 2018). On the other hand, the MiISE5 and MiISE6 effectors from M. incognita, induced changes in the transcriptional regulation of multiple JA signaling genes when overexpressed in A. thaliana (Shi et al. 2018a, b). The MiMsp32, M. incognita effector, also targets the enzyme related to the JA-biosynthesis pathway, which leads to enhancement of the susceptibility of host plants (Verhoeven et al. 2022). Moreover, Meloidogyne effectors are involved in suppressing ROS burst, as for instance, the MgPDI effector from M. graminicola (Tian et al. 2019). Altogether, plant defense mechanisms are frequently a target for nematode effectors, which, as we assume, also occurs in the case of MaMsp4 protein.
Moreover, one of the maize proteins interacting with the MaMsp4 candidate effector and involved in plant defense response is S-adenosyl methionine decarboxylase 2; a key enzyme involved in the biosynthesis of the polyamines: spermidine and spermine (Franceschetti et al. 2001; Gonzalez et al. 2021). Another important enzyme involved in this pathway is spermidine synthase 2, described as a target for H. schachtii effector – 10A06 (Hewezi et al. 2010). Many other pathogens also use phytotoxins and effectors to manipulate plant polyamines biosynthesis pathways, such as Pseudomonas syringae or Ralstonia solanacearum (Gerlin et al. 2021). Polyamines are catabolized by apoplastic oxidases, producing active oxygen species and hydrogen peroxide (H2O2). Moreover, polyamines were described as attractants to M. incognita during the process of identification of suitable host plants (Oota et al. 2020). On the other hand, there is evidence that a silencing suppressor from beet severe curly top virus (BSCTV, Germiniviridae: Curtovirus) manipulates the host’s silencing machinery by targeting S-adenosyl methionine decarboxylase (Guerrero et al. 2020). Furthermore, cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for SA- and leucine-correlated signaling in the defense response to Verticillium dahliae (Mo et al. 2016). In this study, we indicated that the expression of the gene encoding S-adenosyl methionine decarboxylase 2 was suppressed after M. arenaria infection, especially at 7 dpi in root samples.
Another identified MaMsp4-interacting protein, ethanolamine-phosphate cytidylyltransferase, is an essential enzyme in phosphatidylethanolamine synthesis (Song et al. 2022). Phosphatidylethanolamine plays a major role in membrane architecture and is hydrolyzed by phospholipase D, which together with its lipid product play an important role in plant growth, development, and stress response. Additionally, phosphatidylethanolamine is involved in plant immunity (Li and Wang 2019). The gene encoding phospholipase D was induced at the early stage of H. glycines infection of soybean (Alkharouf et al. 2006). Phospholipase is also proposed to be a central regulator in wheat resistance to H. avenae, because of its role in a few different metabolic pathways, activated in the early stage of infection of a resistant host plant (Kong et al. 2015). On the other hand, phosphatidylethanolamines were identified as systemic acquired resistance-related lipid metabolites in plant-bacteria interactions (Song et al. 2022). Moreover, the PldA effector from Pseudomonas aeruginosa achieves its antibacterial activity by degrading phosphatidylethanolamine (Russell et al. 2013). During maize response to M. arenaria infection, the expression level of the gene encoding ethanolamine-phosphate cytidylyltransferase in root samples was down-regulated.
Interestingly, analyses of interacting proteins using the STRING database indicated that ethanolamine-phosphate cytidylyltransferase was found to interact with S-adenosyl methionine, which is a substrate for S-adenosyl methionine decarboxylase, thus, these two MaMsp4 interactors might be functionally connected.
The first group of proteins (beta-galactosidase 11, pectinesterase, and polygalacturonase), known to be involved in cell wall modifications, showed a similar gene coding expression pattern during an infection – they were all overexpressed in the very early stage of infection (24 hpi), while the nematode penetrates the root and then, at a later phase of parasitism (7 dpi), a strong suppression of their expression was observed. It has been observed that for the second group of analyzed proteins (S-adenosyl methionine decarboxylase 2 and ethanolamine-phosphate cytidylyltransferase), which are related to a plant defense response, their gene expression showed down-regulation at 7 dpi, without the preceding overexpression. This result suggests that the MaMsp4 protein might play a role in the suppression of plant defense response, which has been observed for many RKN effectors (Vieira and Gleason 2019). There are also studies suggesting that one effector may play a few roles as for example Mj-FAR-1 protein, secreted by M. javanica (Iberkleid et al. 2015). Authors indicated that this protein was involved in both these processes, cell wall modifications as well as plant defense.
On the basis of the obtained results we have concluded that described in this study M. arenaria protein, MaMsp4, plays an important role during penetration and parasitism in maize and interacts with proteins taking part in plant cell wall modifications as well as with proteins involved in plant defense response mechanisms.
Data availability
Full coding sequence for MaMsp4 candidate effector protein is available in NCBI database under OQ407870 accession number.
Abbreviations
- Ade:
-
Adenine
- Co-IP:
-
Co-immunoprecipitation
- ddPCR:
-
Digital droplet PCR
- DG:
-
Dorsal esophageal gland cells
- dpi:
-
Days post inoculation
- ETI:
-
Effector-triggered immunity
- ETS:
-
Effector-triggered susceptibility
- His:
-
Histidine
- hpi:
-
Hours post inoculation
- JA:
-
Jasmonic acid
- NCBI:
-
National Center for Biotechnology Information
- PAMP:
-
Pathogen-associated molecular pattern
- PRR:
-
Pattern recognition receptor
- PTI:
-
PAMPs-triggered immunity
- RKN:
-
Root-knot nematode
- ROS:
-
Reactive oxygen species
- SA:
-
Salicylic acid
- SvG:
-
Subventral gland cells
- Y2H:
-
Yeast two-hybrid
References
Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224:838–852. https://doi.org/10.1007/s00425-006-0270-8
Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site–leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32:195–204. https://doi.org/10.1046/j.1365-313X.2002.01413.x
Bozbuga R, Lilley CJ, Knox JP, Urwin PE (2018) Host-specific signatures of the cell wall changes induced by the plant parasitic nematode, Meloidogyne incognita. Sci Rep 8:1–13. https://doi.org/10.1038/s41598-018-35529-7
Bryant P, Pozzati G, Elofsson A (2022) Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13:1265. https://doi.org/10.1038/s41467-022-29480-5
Budziszewska M, Frąckowiak P, Obrępalska-Stęplowska A (2021) Analysis of the role of Bradysia impatiens (Diptera: Sciaridae) as a vector transmitting peanut stunt virus on the model plant Nicotiana benthamiana. Cells 10:1546. https://doi.org/10.3390/cells10061546
CABI Compendium (2022) Meloidogyne (root knot nematodes). CABI International. https://doi.org/10.1079/cabicompendium.33231
Chen J, Lin B, Huang Q, Hu L, Zhuo K, Liao J (2017) A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog 13:e1006301. https://doi.org/10.1371/journal.ppat.1006301
Chen J, Hu L, Sun L, Lin B, Huang K, Zhuo K, Liao J (2018) A novel Meloidogyne graminicola effector, MgMO237, interacts with multiple host defence-related proteins to manipulate plant basal immunity and promote parasitism. Mol Plant Pathol 19:1942–1955. https://doi.org/10.1111/mpp.12671
Creasey EA, Delahay RM, Daniell SJ, Frankel G (2003) Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093–2106. https://doi.org/10.1099/mic.0.26355-0
Dinh PT, Brown CR, Elling AA (2014) RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in Arabidopsis and potato. Phytopathology 104:1098–1106. https://doi.org/10.1094/PHYTO-03-14-0063-R
Farine L, Niemann M, Schneider A, Bütikofer P (2015) Phosphatidylethanolamine and phosphatidylcholine biosynthesis by the Kennedy pathway occurs at different sites in Trypanosoma brucei. Sci Rep 5:16787. https://doi.org/10.1074/jbc.M803600200
Franceschetti M, Hanfrey C, Scaramagli S, Torrigiani P, Bagni N, Burtin D, Michael AJ (2001) Characterization of monocot and dicot plant S-adenosyl-l-methionine decarboxylase gene families including identification in the mRNA of a highly conserved pair of upstream overlapping open reading frames. Biochem J 353:403–409. https://doi.org/10.1042/bj3530403
Gao J, Liu Z, Zhao B, Liu P, Zhang J-W (2020) Physiological and comparative proteomic analysis provides new insights into the effects of shade stress in maize (Zea mays L.). BMC Plant Biol 20:1–13. https://doi.org/10.1186/s12870-020-2264-2
Gerlin L, Baroukh C, Genin S (2021) Polyamines: double agents in disease and plant immunity. Trends Plant Sci 26:1061–1071. https://doi.org/10.1016/j.tplants.2021.05.007
Gheysen G, Fenoll C (2002) Gene expression in nematode feeding sites. Annu Rev Phytopathol 40:191–219. https://doi.org/10.1146/annurev.phyto.40.121201.093719
Gleason C, Polzin F, Habash SS, Zhang L, Utermark J, Grundler FM, Elashry A (2017) Identification of two Meloidogyne hapla genes and an investigation of their roles in the plant-nematode interaction. Mol Plant-Microbe Interact 30:101–112. https://doi.org/10.1094/MPMI-06-16-0107-R
Gonzalez ME, Jasso-Robles FI, Flores-Hernández E, Rodríguez-Kessler M, Pieckenstain FL (2021) Current status and perspectives on the role of polyamines in plant immunity. Ann Appl Biol 178:244–255. https://doi.org/10.1111/aab.12670
Goode K, Mitchum MG (2022) Pattern-triggered immunity against root-knot nematode infection: a minireview. Physiol Plant 174:e13680. https://doi.org/10.1111/ppl.13680
Gorshkov V, Tsers I (2022) Plant susceptible responses: the underestimated side of plant–pathogen interactions. Biol Rev 97:45–66. https://doi.org/10.1111/brv.12789
Grossi-de-Sa M, Petitot A-S, Xavier DA, Sá MEL, Mezzalira I, Beneventi MA, Martins NF, Baimey HK, Albuquerque EV, Grossi-de-Sa MF (2019) Rice susceptibility to root-knot nematodes is enhanced by the Meloidogyne incognita MSP18 effector gene. Planta 250:1215–1227. https://doi.org/10.1007/s00425-019-03205-3
Guerrero J, Regedanz E, Lu L, Ruan J, Bisaro DM, Sunter G (2020) Manipulation of the plant host by the geminivirus AC2/C2 protein, a central player in the infection cycle. Front Plant Sci 11:591. https://doi.org/10.3389/fpls.2020.00591
Guo B, Lin B, Huang Q, Li Z, Zhuo K, Liao J (2022) A nematode effector inhibits plant immunity by preventing cytosolic free Ca2+ rise. Plant Cell Environ 45:3070–3085. https://doi.org/10.1111/pce.14406
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
He Y, Karre S, Johal GS, Christensen SA, Balint-Kurti P (2019) A maize polygalacturonase functions as a suppressor of programmed cell death in plants. BMC Plant Biol 19:1–10. https://doi.org/10.1186/s12870-019-1897-5
He P, Zhang J, Lv Z, Cui P, Xu X, George MS, Lu G (2023) Genome-wide identification and expression analysis of the polygalacturonase gene family in sweetpotato. BMC Plant Biol 23:1–13. https://doi.org/10.1186/s12870-023-04272-1
Hewezi T, Howe PJ, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ (2010) Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol 152:968–984. https://doi.org/10.1104/pp.109.150557
Hossain MA (2022) Molecular evolution, three-dimensional structural characteristics, mechanism of action, and functions of plant beta-galactosidases. Bioinformatics in Agriculture. Elsevier, Boston, pp. 191–208. https://doi.org/10.1016/B978-0-323-89778-5.00017-9
Huang G, Gao B, Maier T, Allen R, Davis EL, Baum TJ, Hussey RS (2003) A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Mol Plant-Microbe Interact 16:376–381. https://doi.org/10.1094/MPMI.2003.16.5.376
Hussey R (1973) A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Disease Reports 57:1025–1028
Iberkleid I, Sela N, Brown Miyara S (2015) Meloidogyne javanica fatty acid-and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress-and phenylpropanoid-related genes during nematode infection of tomato. BMC Genomics 16:1–26. https://doi.org/10.1186/s12864-015-1426-3
Ibrahim HM, Hosseini P, Alkharouf NW, Hussein EH, Abd El Kader Y, Aly MA, Matthews BF (2011) Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics 12:1–16. https://doi.org/10.1186/1471-2164-12-220
Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135:1565–1573. https://doi.org/10.1104/pp.104.041699
Jagdale S, Rao U, Giri AP (2021) Effectors of root-knot nematodes: an arsenal for successful parasitism. Front Plant Sci 12:800030. https://doi.org/10.3389/fpls.2021.800030
Jaouannet M, Magliano M, Arguel MJ, Gourgues M, Evangelisti E, Abad P, Rosso M-N (2013) The root-knot nematode calreticulin mi-CRT is a key effector in plant defense suppression. Mol Plant-Microbe Interact 26:97–105. https://doi.org/10.1094/MPMI-05-12-0130-R
Ji H, Gheysen G, Denil S, Lindsey K, Topping JF, Nahar K, Haegeman A, De Vos WH, Trooskens G, Van Criekinge W (2013) Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J Exp Bot 64:3885–3898. https://doi.org/10.1093/jxb/ert219
Jones M (1981) Host cell responses to endoparasitic nematode attack: structure and function of giant cells and syncytia. Ann Appl Biol 97:353–372. https://doi.org/10.1111/j.1744-7348.1981.tb05122.x
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323. https://doi.org/10.1038/nature05286
Jones MG, Goto DB (2011) Root-knot nematodes and giant cells. In: Jones J, Gheysen G, Fenoll C (eds) Genomics and molecular genetics of plant-nematode interactions. Springer, Dordrecht, pp 83–100. https://doi.org/10.1007/978-94-007-0434-3_5
Joshi I, Kumar A, Singh AK, Kohli D, Raman K, Sirohi A, Chaudhury A, Jain PK (2019) Development of nematode resistance in Arabidopsis by HD-RNAi-mediated silencing of the effector gene mi-msp2. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-53485-8
Kamaruzzaman M, Zhao LF, Zhang JA, Zhu LT, Li Y, Deng XD, Cai SJ, Lian XC, Chen FN, Jia N (2023) MiPDCD6 effector suppresses host PAMP-triggered immunity to facilitate Meloidogyne incognita parasitism in tomato. Plant Pathol 72:195–206. https://doi.org/10.1111/ppa.13649
Khan M, Seto D, Subramaniam R, Desveaux D (2018) Oh, the places they'll go! A survey of phytopathogen effectors and their host targets. Plant J 93:651–663. https://doi.org/10.1111/tpj.13780
Kim J, Shiu S-H, Thoma S, Li W-H, Patterson SE (2006) Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol 7:1–14. https://doi.org/10.3390/ijms21165706
Kong L-A, Wu D-Q, Huang W-K, Peng H, Wang G-F, Cui J-K, Liu S-M, Li Z-G, Yang J, Peng D-L (2015) Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics 16:1–18. https://doi.org/10.1186/s12864-015-2037-8
Kumar A, Fitoussi N, Sanadhya P, Sichov N, Bucki P, Bornstein M, Belausuv E, Brown Miyara S (2023) Two candidate Meloidogyne javanica effector genes, MjShKT and MjPUT3: a functional investigation of their roles in regulating nematode parasitism. Mol Plant-Microbe Interact 36:79–94. https://doi.org/10.1094/MPMI-10-22-0212-R
Leelarasamee N, Zhang L, Gleason C (2018) The root-knot nematode effector MiPFN3 disrupts plant actin filaments and promotes parasitism. PLoS Pathog 14:e1006947. https://doi.org/10.1371/journal.ppat.1006947
Li J, Wang X (2019) Phospholipase D and phosphatidic acid in plant immunity. Plant Sci 279:45–50. https://doi.org/10.1016/j.plantsci.2018.05.021
Lin B, Zhuo K, Wu P, Cui R, Zhang L-H, Liao J (2013) A novel effector protein, MJ-NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol Plant-Microbe Interact 26:55–66. https://doi.org/10.1094/MPMI-05-12-0114-FI
Lin B, Zhuo K, Chen S, Hu L, Sun L, Wang X, Zhang LH, Liao J (2016) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol 209:1159–1173. https://doi.org/10.1111/nph.13701
Long H, Wang X, Xu J (2006) Molecular cloning and life-stage expression pattern of a new chorismate mutase gene from the root-knot nematode Meloidogyne arenaria. Plant Pathol 55:559–563. https://doi.org/10.1111/j.1365-3059.2006.01362.x
Lorrai R, Ferrari S (2021) Host cell wall damage during pathogen infection: mechanisms of perception and role in plant-pathogen interactions. Plants 10:399. https://doi.org/10.3390/plants10020399
Lyska D, Engelmann K, Meierhoff K, Westhoff P (2013) pAUL: a gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS One 8:e53787. https://doi.org/10.1371/journal.pone.0053787
Mahalingam R, Wang G, Knap HT (1999) Polygalacturonase and polygalacturonase inhibitor protein: gene isolation and transcription in Glycine max-Heterodera glycines interactions. Mol Plant-Microbe Interact 12:490–498. https://doi.org/10.1094/MPMI.1999.12.6.490
Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5:178. https://doi.org/10.3389/fpls.2014.00178
Mani V, Reddy CS, Lee S-K, Park S, Ko H-R, Kim D-G, Hahn B-S (2020) Chitin biosynthesis inhibition of Meloidogyne incognita by RNAi-mediated gene silencing increases resistance to transgenic tobacco plants. Int J Mol Sci 21:6626. https://doi.org/10.3390/ijms21186626
Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815. https://doi.org/10.1016/j.jplph.2012.01.019
Mejias J, Truong NM, Abad P, Favery B, Quentin M (2019) Plant proteins and processes targeted by parasitic nematode effectors. Front Plant Sci 10:970. https://doi.org/10.3389/fpls.2019.00970
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben M, Davis EL (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol 199:879–894. https://doi.org/10.1111/nph.12323
Mo H-J, Sun Y-X, Zhu X-L, Wang X-F, Zhang Y, Yang J, Yan G-J, Ma Z-Y (2016) Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid-and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 243:1023–1039. https://doi.org/10.1007/s00425-015-2463-5
Moreira VJV, Lourenço-Tessutti IT, Basso MF, Lisei-de-Sa ME, Morgante CV, Paes-de-Melo B, Arraes FBM, Martins-de-Sa D, Silva MCM, de Almeida EJ (2022) Minc03328 effector gene downregulation severely affects Meloidogyne incognita parasitism in transgenic Arabidopsis thaliana. Planta 255:44. https://doi.org/10.1007/s00425-022-03823-4
Naalden D, Haegeman A, de Almeida-Engler J, Birhane Eshetu F, Bauters L, Gheysen G (2018) The Meloidogyne graminicola effector Mg16820 is secreted in the apoplast and cytoplasm to suppress plant host defense responses. Mol Plant Pathol 19:2416–2430. https://doi.org/10.1111/mpp.12719
Ngou BPM, Ding P, Jones JD (2022) Thirty years of resistance: zig-Zag through the plant immune system. Plant Cell 34:1447–1478. https://doi.org/10.1093/plcell/koac041
Nguyen C-N, Perfus-Barbeoch L, Quentin M, Zhao J, Magliano M, Marteu N, Da Rocha M, Nottet N, Abad P, Favery B (2018) A root-knot nematode small glycine and cysteine-rich secreted effector, MiSGCR1, is involved in plant parasitism. New Phytol 217:687–699. https://doi.org/10.1111/nph.14837
Niu J, Liu P, Liu Q, Chen C, Guo Q, Yin J, Yang G, Jian H (2016) Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci Rep 6:19443. https://doi.org/10.1038/srep19443
Oota M, Tsai AY-L, Aoki D, Matsushita Y, Toyoda S, Fukushima K, Saeki K, Toda K, Perfus-Barbeoch L, Favery B (2020) Identification of naturally occurring polyamines as root-knot nematode attractants. Mol Plant 13:658–665. https://doi.org/10.1016/j.molp.2019.12.010
Passianotto ALL, Sonah H, Dias WP, Marcelino-Guimarães FC, Belzile F, Abdelnoor RV (2017) Genome-wide association study for resistance to the southern root-knot nematode (Meloidogyne incognita) in soybean. Mol Breed 37:1–11. https://doi.org/10.1007/s11032-017-0744-3
Petersen TN, Brunak S, Von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Petitot A-S, Dereeper A, Da Silva C, Guy J, Fernandez D (2020) Analyses of the root-knot nematode (Meloidogyne graminicola) transcriptome during host infection highlight specific gene expression profiling in resistant rice plants. Pathogens 9:644. https://doi.org/10.3390/pathogens9080644
Pham A-T, McNally K, Abdel-Haleem H, Roger Boerma H, Li Z (2013) Fine mapping and identification of candidate genes controlling the resistance to southern root-knot nematode in PI 96354. Theor Appl Genet 126:1825–1838. https://doi.org/10.1007/s00122-013-2095-8
Przybylska A, Obrępalska-Stęplowska A (2020) Plant defense responses in monocotyledonous and dicotyledonous host plants during root-knot nematode infection. Plant Soil 451:239–260. https://doi.org/10.1007/s11104-020-04533-0
Przybylska A, Spychalski M (2021) Changes in the expression level of genes encoding transcription factors and cell wall-related proteins during Meloidogyne arenaria infection of maize (Zea mays). Mol Biol Rep 48:6779–6786. https://doi.org/10.1007/s11033-021-06677-3
Przybylska A, Kornobis F, Obrępalska-Stęplowska A (2018) Analysis of defense gene expression changes in susceptible and tolerant cultivars of maize (Zea mays) upon Meloidogyne arenaria infection. Physiol Mol Plant Pathol 103:78–83. https://doi.org/10.1016/j.pmpp.2018.05.005
Pu W, Xiao K, Luo S, Zhu H, Yuan Z, Gao C, Hu J (2022) Characterization of five Meloidogyne incognita effectors associated with PsoRPM3. International J Mol Sci 23:1498. https://doi.org/10.3390/ijms23031498
Qiao F, Kong L-A, Peng H, Huang W-K, Wu D-Q, Liu S-M, Clarke JL, Qiu D-W, Peng D-L (2019) Transcriptional profiling of wheat (Triticum aestivum L.) during a compatible interaction with the cereal cyst nematode Heterodera avenae. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-018-37824-9
Ridgway ND (2021) Phospholipid synthesis in mammalian cells. In: Ridgway ND, McLeod RS (eds) Biochemistry of lipids, lipoproteins and membranes. Elsevier, Boston, pp 209–236. https://doi.org/10.1016/B978-0-444-63438-2.00007-9
Rocha R, Hussey R, Pepi L, Azadi P, Mitchum M (2023) Discovery of novel effector protein candidates produced in the dorsal gland of root-knot nematode adult females. Mol Plant-Microbe Interact. https://doi.org/10.1094/MPMI-11-22-0232-R
Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD (2013) Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. https://doi.org/10.1038/nature12074
Seddigh S, Darabi M (2014) Comprehensive analysis of beta-galactosidase protein in plants based on Arabidopsis thaliana. Turk J Biol 38:140–150. https://doi.org/10.3906/biy-1307-14
Shi Q, Mao Z, Zhang X, Ling J, Lin R, Zhang X, Liu R, Wang Y, Yang Y, Cheng X, Xie B (2018a) The novel secreted Meloidogyne incognita effector MiISE6 targets the host nucleus and facilitates parasitism in Arabidopsis. Front Plant Sci 9. https://doi.org/10.3389/fpls.2018.00252
Shi Q, Mao Z, Zhang X, Zhang X, Wang Y, Ling J, Lin R, Li D, Kang X, Sun W (2018b) A Meloidogyne incognita effector MiISE5 suppresses programmed cell death to promote parasitism in host plant. Sci Rep 8:7256. https://doi.org/10.1038/s41598-018-24999-4
Sobczak M, Fudali S, Wieczorek K (2011) Cell wall modifications induced by nematodes. In: Jones J, Gheysen G, Fenoll C (eds) Genomics and molecular genetics of plant-nematode interactions. Springer, Dordrecht, pp 395–422. https://doi.org/10.1007/978-94-007-0434-3_19
Song H, Lin B, Huang Q, Sun L, Chen J, Hu L, Zhuo K, Liao J (2021) The Meloidogyne graminicola effector MgMO289 targets a novel rice copper metallochaperone to suppress plant immunity. J Exp Bot 72(15):5638–5655. https://doi.org/10.1093/jxb/erab208
Song J-B, Huang R-K, Guo M-J, Zhou Q, Guo R, Zhang S-Y, Yao J-W, Bai Y-N, Huang X (2022) Lipids associated with plant-bacteria interaction identified using a metabolomics approach in an Arabidopsis thaliana model. PeerJ 10:e13293. https://doi.org/10.7717/peerj.13293
Steinegger M, Söding J (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35:1026–1028. https://doi.org/10.1038/nbt.3988
Stojilkovic B, Xiang H, Chen Y, Bauters L, Van de Put H, Steppe K, Liao J, de Almeida Engler J, Gheysen G (2022) The nematode effector Mj-NEROSs interacts with ISP influencing plastid ROS production to suppress plant immunity. bioRxiv: 2022.2010. 2024.513376. https://doi.org/10.1101/2022.10.24.513376
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Thordal-Christensen H, Birch PR, Spanu PD, Panstruga R (2018) Why did filamentous plant pathogens evolve the potential to secrete hundreds of effectors to enable disease? Mol Plant Pathol 19:781–785. https://doi.org/10.1111/mpp.12649
Tian Z-l, Wang Z-h, Maria M, Qu N, Zheng J-w (2019) Meloidogyne graminicola protein disulfide isomerase may be a nematode effector and is involved in protection against oxidative damage. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-48474-w
Verhoeven A, Finkers-Tomczak A, Prins P, Valkenburg-van Raaij DR, van Schaik CC, Overmars H, van Steenbrugge JJ, Tacken W, Varossieau K, Slootweg EJ (2022) The root-knot nematode effector MiMSP32 targets host 12-oxophytodienoate reductase 2 (OPR2) to regulate plant susceptibility. New Phytol 237(6):2360–2374. https://doi.org/10.1111/nph.18653
Veronico P, Rosso LC, Melillo MT, Fanelli E, De Luca F, Ciancio A, Colagiero M, Pentimone I (2022) Water stress differentially modulates the expression of tomato cell wall metabolism-related genes in Meloidogyne incognita feeding sites. Front Plant Sci 13:776. https://doi.org/10.3389/fpls.2022.817185
Vieira P, Gleason C (2019) Plant-parasitic nematode effectors—insights into their diversity and new tools for their identification. Curr Opin Plant Biol 50:37–43. https://doi.org/10.1016/j.pbi.2019.02.007
Wang X, Xue B, Dai J, Qin X, Liu L, Chi Y, Jones J, Li H (2018) A novel Meloidogyne incognita chorismate mutase effector suppresses plant immunity by manipulating the salicylic acid pathway and functions mainly during the early stages of nematode parasitism. Plant Pathol 67:1436–1448. https://doi.org/10.1111/ppa.12841
Zhao J, Mao Z, Sun Q, Liu Q, Jian H, Xie B (2020) MiMIF-2 effector of Meloidogyne incognita exhibited enzyme activities and potential roles in plant salicylic acid synthesis. Int JMol Sci 21:3507. https://doi.org/10.3390/ijms21103507
Zhou J-j, Zhang X-p, Rui L, Jian L, Yan L, Yang Y-h, Xie B-y, Zhao J-l, Mao Z-c (2023) A Meloidogyne incognita effector Minc03329 suppresses plant immunity and promotes parasitism. J Integr Agric 22:799–811. https://doi.org/10.1016/j.jia.2022.08.117
Zhuo K, Chen J, Lin B, Wang J, Sun F, Hu L, Liao J (2017) A novel Meloidogyne enterolobii effector MeTCTP promotes parasitism by suppressing programmed cell death in host plants. Mol Plant Pathol 18:45–54. https://doi.org/10.1111/mpp.12374
Zhuo K, Naalden D, Nowak S, Xuan Huy N, Bauters L, Gheysen G (2019) A Meloidogyne graminicola C-type lectin, Mg01965, is secreted into the host apoplast to suppress plant defence and promote parasitism. Mol Plant Pathol 20:346–355. https://doi.org/10.1111/mpp.12759
Acknowledgments
We would like to thank D. Baulcombe (University of Cambridge, Cambridge, UK) for providing a pBin61 vector and K. Meierhoff (Heinrich-Heine-Universitaet, Duesseldorf, Germany) for providing a pAUL1 vector. The A. tumefaciens strain EHA105 was kindly gifted from T. Pniewski (Institute of Plant Genetics Polish Academy of Science, Poznań, Poland). We also would like to thank John Jones (James Hutton Institute, Dundee, Scotland) and Sophie Mantelin (French National Institute for Agriculture, Food, and Environment, Sophia Antipolis, France) for help with a potential M. arenaria candidate effector determination and technical advice.
Funding
This study was supported by a grant from the Polish National Science Center [grant number 2014/13/N/NZ9/00703] and by a project from the Ministry of Education and Science [BIOTECH-01].
Author information
Authors and Affiliations
Contributions
Arnika Przybylska and Aleksandra Obrępalska-Stęplowska contributed to the study conception and design. Material preparation, data collection, and experiments were carried out by Arnika Przybylska and Przemysław Wieczorek. All authors analysed and interpreted the results. The first draft of the manuscript was written by Arnika Przybylska and Aleksandra Obrępalska-Stęplowska and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Tida Ge.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Przybylska, A., Wieczorek, P. & Obrępalska-Stęplowska, A. Meloidogyne arenaria candidate effector MaMsp4 interacts with maize (Zea mays L.) proteins involved in host defense response and cell wall modifications. Plant Soil 491, 501–523 (2023). https://doi.org/10.1007/s11104-023-06130-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06130-3