Abstract
Background
Sustainable agriculture seeks to optimize the application of nitrogen (N) fertilizers to reduce adverse economic and ecological effects. Crop diversification has been proposed to increase the efficiency of N fertilization. An open question is how the soil microbiome responds to these beneficial practices.
Methods
In a field study we investigated the effects of mineral N fertilizer with a 0-control, a conventional amount of 150 kg N ha−1 and an excessive application of 250 kg N ha−1 on the soil microbiome within a diversified cropping system with oil radish and undersown ryegrass over a period of 2.5 years and a non-diversified control, both in rotation of potato, winter rye and maize.
Results
N-fertilizations and crop rotations altered the pH, but differences were less pronounced with the diversified system. Compared to the crop species and season, N fertilization and crop diversification had less influence on the abundance of soil bacteria, archaea and fungi. The crop diversification showed a much stronger effect on archaeal than on bacterial or fungal abundances, while the microbial carbon use efficiency correlated strongly with bacterial abundance. At the end of the growing seasons, crop diversification increased prokaryotic richness and Shannon diversity in response to N addition, with a greater increase in the conventional N. At conventional N supply, prokaryotic co-occurrence networks revealed a much denser and complex structure in the diversified system.
Conclusions
The diversified cropping system under conventional N application rates showed positive effects on the prokaryotic soil microbiome by increasing their richness, Shannon diversity, and promoting a more elaborated network structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N) is a major growth limiting factor in cropping systems and has therefore traditionally been applied in agricultural systems in plentiful amounts as organic or mineral N-fertilizers (Foyer et al. 2016; Vitousek et al. 2002). Such surplus N applications however are not only economically wasteful but may also trigger environmental problems. More than half of applied N fertilizer can be lost from agricultural fields due to nitrate leaching and nitrogenous oxide gas emission, both potentially also resulting from microbial N transformations (Erisman et al. 2008; Foyer et al. 2016; Francioli et al. 2016; Hartmann and Six 2022; He et al. 2020).
Nitrate leaching from agricultural soils can be greatly affected by the employed cropping system (Beaudoin et al. 2005; Hall et al. 2001; Smith et al. 2013; Tonitto et al. 2006; Toth and Fox 1998). Inclusion of cover crops into the crop rotation as a diversification strategy has been shown to improve N use efficiencies of plants (Hauggaard-Nielsen et al. 2012; Kankanen and Eriksson 2007; Thorup-Kristensen et al. 2012). Introduction of undersown ryegrass in cereal/wheat production systems reduced N leaching by 50%, as shown by a meta-analysis of 35 studies in Nordic countries (Valkama et al. 2015). While such cases have demonstrated the usefulness of diversification in increasing N-use efficiency of crops, there is still limited understanding of the molecular mechanisms underlying the role of diversified cropping systems in limiting N losses.
It is well known that the soil microbiome carries out all crucial steps in the biogeochemical cycling of N, including nitrification, denitrification, N fixation, or the nitrite-dependent oxidation of ammonium (Kuypers et al. 2018). A diversified cropping system could prevent the N losses by supporting the ecosystem function of the soil microbial community in soil biogeochemical cycling, e.g. via building up more biomass and stabilizing a more diversified microbiome, since the diversity of the microbial community correlates with its capacity to transform N (Delgado-Baquerizo et al. 2016). In agricultural fields, diversified cropping systems, like cover crops were shown to improve soil structure by the formation of aggregates, which provide diverse microhabitats, likely to promote a more diverse microbiome (Qi et al. 2022). It also could increase the soil microbial biomass carbon (C) and N by at least 40% as revealed from 81 different studies (Muhammad et al. 2021). Furthermore, an analysis of 60 studies found that soil microbial abundance and activity increased with cover crops by up to 27%, (Kim et al. 2020). In tropical cropping systems with maize it was demonstrated that the presence of cover crops affected the abundance of microbial N-cycling genes, and thereby possibly increase N-fertilization efficiency (Momesso et al. 2022; Rocha et al. 2020), and this may also apply to other climatic conditions where crops are cultivated. Given the complex composition and high diversity of the microbiome, including bacteria, archaea and fungi, with their specific and highly heterogenous properties, different soil microbial taxa may not equally respond to crop diversification and change of N supply e.g., cover crops favored fungi more than bacteria, and ammonia-oxidizing archaea showed a stronger correlation with cover crop biomass than ammonia-oxidizing bacteria (Momesso et al. 2022; Muhammad et al. 2021). While these studies demonstrate that microbial groups and functional potentials can differentially be affected by specific cropping systems, it is still difficult to see a more general response pattern of the soil microbiome including those mediating N transformation processes.
Microbial carbon use efficiency (CUE) is a physiological parameter that indicates how energy rich carbon resources are utilized for microbial growth versus immediate loss by maintenance respiration (Geyer et al. 2019). Its usefulness to describe the response of the soil microbiome to N fertilization was already explored but the influence of cropping systems is still poorly understood. N fertilization could reduce microbial C uptake with lower microbial respiration and lower microbial growth rate, potentially by inhibiting the degradation of aromatic compounds (Spohn et al. 2016). However, studies with grassland microbiomes found CUE to be robust towards changes imposed by N and phosphate fertilization (Widdig et al. 2020). Such contrasting results of CUE could be an outcome of both direct and indirect effects on the microhabitat conditions e.g., in agricultural systems rhizodeposition is an important factor that can influence microbial habitats (Denef et al. 2009). Microorganisms tended to use more root-derived C and presented higher microbial CUE in the rhizosphere with abundant N fertilization, potentially owing to the low energy costs for cell uptake under the condition of high C supply, which was associated with higher N fertilizations (Bicharanloo et al. 2020). In fact, most studies on CUE in cropping systems suffer from the fact that only single sampling events were analyzed and such snapshots may not sufficiently consider the impact of seasonal changes and crop rotations over years. Both have a tremendous effect on the living conditions of soil microorganisms (Liu et al. 2022; Simon et al. 2020). Thus, we still lack a more systematic understanding of the impacts of mineral N fertilization and crop diversification on soil microbial CUE, specific microbial functional groups and the temporal resilience of compositional changes of the soil microbiome.
The objective of this study was to elucidate the effects of different mineral N fertilization rates and the influence of crop diversification on soil physicochemical parameters, soil microbial CUE and the diversity of the soil microbiome, with emphasis to their prokaryotic community members. From an agricultural field experiment located in Lower Saxony, Germany, we collected soil samples over a period of 2.5 years before and after the cropping season from two cropping systems (winter cover crop and undersown ryegrass vs. non-diversified) amended with an excessive amount of 250 kg N ha−1 (250N), a conventional amount of 150 N and a control without any additional N supply. The dynamics of physicochemical soil parameters (pH, Corg and Ntotal) were followed along with a cropping regime including a succession of potato, winter rye and maize. The determination of soil microbial CUE was conducted on the soil samples collected from the first three sampling events, while the prokaryotic community dynamics were investigated in those soil samples from the first four sampling events. We hypothesized that conventional 150 N fertilization combined with crop diversification will increase the soil microbial diversity and microbial CUE by providing plentiful C and N sources. In contrast, we suspected that the excessive N fertilization would decrease microbial diversity as well as their CUE by accelerating soil acidification and thereby neutralize the positive effect of crop diversification seen at lower N supplies. Finally, because of the different microhabitat conditions and nutrient supplies in spring and autumn, we assumed that soil microbiomes as analyzed before and after the cropping season would show seasonal differences.
Materials and methods
Experimental site and soil sampling
The soil was collected from an ongoing crop diversification and N fertilization trial initiated in 2014, located at Hamerstorf, Germany (52°54′22″ N, 10°27′6″ E, 50 m a.s.l.). This field site is maintained as part of a field-experimental-system under the technical authority of the Chamber of Agriculture Lower Saxony (Landwirtschaftskammer Niedersachsen), and it is financed by the water-treatment charge of Lower Saxony. The average annual precipitations of the region were 406 mm, 628 mm and 542 mm, while the mean annual temperatures were 10.4 °C, 10.4 °C and 10.6 °C from 2018 to 2020, respectively. The soil type was classified as silty sand (Su2) with 83% sand, 14% silt, and 3% clay, respectively.
This experimental field simulated a model for a water protection area to analyze how risks of N leaching into ground and surface water can be reduced by crop diversification and N fertilizer management. There were two cropping trials in a split-block design, including a control system and crop diversification system, both cultivated with potato (Solanum tuberosum), winter rye (Secale cereale), and maize (Zea mays) from 2018 to 2020, respectively (Fig. 1). The diversified cropping system included winter cover crops (oil radish) between winter rye (2019) and potato (2018) cultivations, and undersown ryegrass with cultivation of winter rye and maize. In addition to the crop diversification, different amounts of mineral N fertilizer treatments were implemented in each cropping trial. The mineral N fertilizer consisted of half ammonium and half nitrate nitrogen. Irrespective of the treatments, above ground crop residues were removed after harvesting. This study selected three N fertilization treatments including T1 (without N fertilization, abbreviated here as 0N), T4 (amended with a conventional amount of 150 kg N ha−1; 150N) and T6 (amended with an excessive amount of 250 kg N ha−1; 250N). Prior to this study during the growing season of 2018 and adjusted for the N demand of potato, the conventional amount of N fertilizer was 160 kg N ha−1 in T4 treatment, and 240 kg N ha−1 in T6 treatment, respectively. Each treatment included four replicate plots ordered in a randomized block design (9 m × 12 m), which were all sampled. The fertilizers were applied at the beginning and the middle of the respective crop growth cycles.
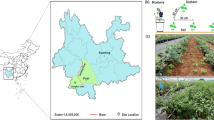
a Aerial view of the Hamerstorf field site indicating the field blocks with control and diversified cropping systems (image generated by Google Earth on June 2, 2017); b Schematic view of the field plots with their respective N applications; c Crop rotations of the two cropping systems (control and crop diversification) since 2015. The red lined frame indicates the time period of sampling for this study
Soil samples were collected in spring (S) at the onset and in autumn (A) at the end of the growing season. The exact sampling dates mainly depended on the weather condition and local agronomic plan, thus it varied between years. The sampling date in autumn 2018 was October 25 (abbreviated here as 2018A), in spring 2019 March 25 (2019S) and in autumn October 24 (2019A), in spring 2020 May 7 (2020S) and in autumn October 15 (2020A). Soil samples were collected by shovel and sampled from four spatially distinct spots in the center area of each plot from a depth of 1 to 10 cm after gently removing the upper-most surface soil. Soils from these spots were well mixed to generate a composite sample of each plot. These soil samples were kept at ambient temperature for a maximum of 3 h and then processed in the laboratory, where the soil was first air-dried and then sieved to 2 mm in order to remove residual straw and plant roots. Soil pH was measured in 0.01 M calcium chloride with a pH meter (HI221 Microprocessor, Hanna Instruments Germany). Corg and Ntotal were analyzed by dry combustion in an elemental analyzer (Leco TruMac, St. Joseph, MI). Soil pH, Corg and Ntotal were measured for all soil samples collected in this study, and so was the determination of the abundance of bacteria, archaea and fungi by quantitative PCR (qPCR). Prokaryotic diversity and community composition were characterized from samples of the first four sampling events (2018A to 2020S), thus with annual replication for each season, and soil microbial carbon use efficiency (CUE) was determined from samples of the first three sampling events. Analyses of these parameters were always based on all four independent replicates of each treatment.
DNA extraction, quantification and sequencing
Microbial DNA was extracted from 500 mg of soil with the FastDNA®SPIN Kit (MP Biomedicals, Eschwege, Germany) following the manufacturer’s protocol. The DNA samples were stored at -20° C for gene quantification and -80° C for 16S rRNA sequencing. The abundance of bacteria, archaea and fungi was quantified with the qPCR method by using a Biorad CFX96 Realtime PCR cycler with C1000 Touch (Biorad, Feldkirchen, Germany). TaqMan assays were used to estimate bacterial 16S rRNA genes using the primer pair BAC338F/BAC805R with probe BAC516F (Yu et al. 2005). The archaeal gene copy numbers were quantified with primers and probe: ARC787F, ARC1059R and ARC915F (Yu et al. 2005). The qPCR reactions for bacteria and archaea were conducted in 20 µL reactions containing 10 µL SYBR (Maxima SYBR Green qPCR, Master Mix 2X no ROX, Thermo Fisher Scientific, Erlangen, Germany), 0.2 µL of each primer (50 µM), 0.08 µL of 50 µM TaqMan, 7.52 µL of DNA/RNAase-free water for qPCR (Thermo Fisher Scientific) and 2 µL of extracted soil DNA. The qPCR cycling conditions for bacterial and archaeal gene quantification consisted of an initial 10 min at 95 °C followed by 39 cycles of denaturing at 95 °C for 15 s and annealing/polymerase extension at 60 °C for 1 min. The abundance of fungi was estimated by quantifying a fragment of the ITS1 region by qPCR, using the primer pair NSII/ 58A2R, in 20 µL reactions: 10 µL SYBR (Maxima SYBR Green qPCR, Master Mix 2 X no ROX, Thermo Fisher Scientific), 0.2 µL of each primer (50 µM), 7.6 µL of DNA/RNAase-free water for qPCR (Thermo Fisher Scientific) and 2 µL of extracted soil DNA (Martin and Rygiewicz 2005). Its cycling condition consisted of an initial 10 min at 95 °C, followed by 39 cycles of 95 °C for 15 s, annealing at 52 °C for 30 s, extension at 72 °C for 30 s and 79 °C for 15 s. Standard curves for bacteria, archaea and fungi were prepared with culture strains obtained from the DSMZ, Braunschweig, Germany, i.e. Bacillus subtilis, Methanobacterium oryzae (DSM 11,106) and Fusarium culmorum (DSM 62,191), respectively. Amplification efficiencies were 95% (bacteria), 96% (archaea) and 93% (fungi); correlation coefficients were above 0.998 for bacteria, 0.996 for archaea, and 0.995 for fungi.
For 16S rRNA sequencing, the V4 region of the 16S rRNA gene was amplified with the 515F/806R primer set (Caporaso et al. 2011) and sequenced as described elsewhere (Herbold et al. 2015). Paired-end sequencing (2 × 300 bp) for the 16S rRNA gene PCR amplicons was performed on the Illumina MiSeq platform by LGC Genomic GmbH, Berlin, Germany. All the samples were sequenced together in the same run and all the sequencing data were analyzed together with the QIIME2 platform (Bolyen et al. 2019). Considering the blind ligation of Illumina adaptor and amplicon fragments, raw sequence reads were firstly reoriented into respective forward and reverse read files by using an in-house Python script (github.com/DamienFinn/MiSeq_read_reorientation). The Cutadapt paired-end method was used to demultiplex the reoriented sequence reads on the QIIME2 platform (Martin 2011). Later, the forward and reverse reads were merged by Vsearch join-pairs function (Rognes et al. 2016). After the merge step, sequence reads were truncated at positions 280 and 40 by DADA2 denoise-single function (Callahan et al. 2016). In this study, amplicon sequence variants (ASV) were adopted and assigned with the Silva 138 database (Quast et al. 2013; Yilmaz et al. 2014). Eukaryote-associated ASV (mitochondria and chloroplasts) were removed. The DNA sequence reads have been deposited in the European Nucleotide Archive database https://www.ebi.ac.uk/ena/browser/ Project Accession number PRJEB47841).
Determination of soil microbial growth, respiration and carbon use efficiency (CUE)
Soil microbial CUE was determined with the 18O-labeling method. This method quantifies microbial growth via the incorporation of 18O from H218O into microbial DNA as previously described in detail (Poeplau et al. 2019; Schroeder et al. 2021). Briefly, 20 g to 25 g fresh weight of soil from each sample was adjusted to 45% (wt/wt) water-holding capacity, then preincubated for 7 d at 15 °C in the dark. After the preincubation, two aliquots of approximately 400 mg fresh weight of soil were collected into a 2 mL incubation microtube and put into a 20 mL air-tight gas vial later sealed with a rubber and aluminum cap for the assessment of 18O-incorporation into newly formed DNA, where one aliquot represented the natural abundance reference and the other was labelled. The soil water content in the two incubation microtubes per sample was adjusted to 60% water holding capacity with ddH2O in the unlabeled incubation microtube and with H218O (97% atom percent w/w) in labelled incubation microtube with the syringe. Specifically, the 18O content in the final soil water was maintained at 20% (wt/wt). In order to allow the determination of microbial respiration rates from the increase in CO2 concentration, the starting conditions of the labeled samples were adjusted to 349 ppm CO2 and 1.3 bar. Both unlabeled and labeled incubation microtubes were kept at 15 °C for 24 h in the dark. Headspace gas samples were extracted only from the labelled incubation microtubes using a gas-tight syringe for CO2 concentration measurement to determine the respiration rate, while the soil samples from both unlabeled and labeled incubation microtubes were immediately frozen with liquid N2 and stored at -80 °C for DNA extraction. The CO2 concentration was measured by gas chromatography with an electron capture detector (Agilent 7890A GC, Agilent Technologies). The amount of respired CO2 (microbial respiration, ng C g−1 dry weight soil h−1) was calculated as described before (Schroeder et al. 2021). DNA was extracted by FastDNA®−SPIN Kit (MP Biomedicals, Eschwege, Germany) and eluted in 100 μL DNase-free water. Later the DNA concentration was quantified by QuantiT PicoGreen dsDNA Kit (Invitrogen, Carlsbad. CA, USA). And 60 μL DNA from both unlabeled and labeled samples were dried at 60 °C in silver capsules. The abundance of 18O was measured by a high-temperature conversion/elemental analyzer (TC/EA) (Thermo Fisher Scientific) coupled with a Delta V Plus isotope ratio mass spectrometer via a ConFloIV interface (Thermo Fisher Scientific). The total DNA produced in 24 h (DNAproduced, μg) was calculated as described in detail elsewhere (Poeplau et al. 2019; Schroeder et al. 2021).
To convert newly formed DNA into C directed to microbial biomass during the 24-h incubation, a conversion factor is needed (Spohn et al. 2016; Schroeder et al. 2021). Therefore, microbial biomass was measured on 7 g dry mass aliquots of soil using the chloroform-fumigation extraction method (Vance et al. 1987). The fumigation process lasted 24 h at room temperature in a dark environment. Fumigated and non-fumigated samples were extracted with 0.5 M K2SO4 solution in a 1:4 dry weight/volume ratio. The Corg was measured in a DIMATOC 2000 (Dimatec, Essen, Germany). The efficiency of microbial C extraction was assumed as 0.45 (Joergensen 1996). Based on the result of microbial biomass C content (μg C g−1 dw soil) and DNA concentration (μg g−1 dw soil) derived from PicoGreen screening, the conversion factor fDNA was determined by their ratio for each sample. This conversion factor fDNA was used to calculate the amount of microbial biomass C production (microbial growth, ng C g−1 dw soil h−1) during 24 h incubation based on the DNAproduced (μg) derived from 18O labelling for each sample. The CUE was indicated by the ratio of microbial biomass C production (microbial growth) and total C uptake, which was the sum of microbial respiration and microbial growth.
Data analysis
Rarefaction analysis was performed to estimate the sequence coverage with the vegan package (v.2.5–7) in R (v.4.0.3) (Revelle 2022). Sequences per sample were rarefied to 6,311 to normalize sequencing depth for the following analyses. The Shannon index, richness and evenness were calculated in R using the vegan package. Fisher’s Least Significant Difference (LSD) post hoc test was performed with the agricolae package (v. 1.3–3) in R (Mendiburu 2021), and the significant differences were considered as p < 0.05. The fit of LMEs was calculated with restricted maximum likelihood with the lmerTest package (v. 3.1–3) in R (Kuznetsova et al. 2017). Spearman’s correlation coefficient was calculated with the package stats (v. 4.1.1). Non-metric multidimensional scaling (NMDS) and Analysis of similarities (ANOSIM) tests were conducted with the vegan package (v.2.5–7). Statistically significant differences in taxonomic ranks (family and ASV) between different treatments were calculated using the Aldex2 package (v.1.22.0) in R, and the significant differences were considered as Benjamini-Hochberg-corrected P-values less than 0.05 (Fernandes et al. 2013, 2014). Network analysis was performed to explore the microbial co-occurrence patterns with the psych package (v.2.1.9), funrar package (v.1.4.1) and igraph package (v.1.2.6) in R (Csardi and Nepusz 2006; Grenié et al. 2017; Revelle 2022; Wickham et al. 2021). For generating correlation networks only ASV with an average read of above one in all samples were considered. More stringent conditions with higher average reads of above 3, 5 and 7 were also analyzed. The correlation matrix was generated by the psych package, and correlation coefficients (r > 0.7 or r < − 0.7) and FDR (false discovery rate) corrected p-values less than 0.05 were used to form the networks. The network images were created with igraph package, as well as the topological features. The microbial functional profile was predicted using the Functional Annotation of Prokaryotic Taxa (FAPROTAX, v.1.2.4) (Louca et al. 2016). FAPROTAX is a database which can map taxonomic annotation profiles of ASV to their potential functions. It is a software for converting ASV into putative functional profiles. It distinguishes over 80 different functions and considers over 7,500 functional annotations obtained from over 4,600 taxa.
Results
Effect of N application rates and crop diversification on physicochemical soil parameters
Both mineral N fertilization and cropping regimes significantly affected the soil pH (p < 0.05; Table 1). The acidification effect of N addition was significant (p < 0.01) at all five sampling events, and, for the non-diversified cropping system, the shift was related to the amount of N added. In the winter rye season, the crop growth increased the soil pH by 0.7 in 150N and 0.8 in 250N on average while maize cultivation caused a decline in soil pH value by 0.6 in 150N and 0.9 in 250N on average (p < 0.05). In contrast, for the diversified system, there was no clear relationship with the amount of N added. The pH shift after winter rye crop was only around 0.1 on average with both 150N and 250N, respectively. Similarly, maize cultivation caused a relatively minor decline in soil pH around 0.4, irrespective of the amount of N supply.
Neither mineral N fertilization nor crop diversification affected the soil Corg (p > 0.05). In contrast, cultivation of winter rye appeared to increase Corg, especially in diversified cropping systems, while maize cultivation caused a slight reduction, which was on average higher with 2.56% in non-diversified cropping systems than in diversified cropping systems with 1.86% (p < 0.05). Irrespective of diversification or N supply, Corg showed a clear trend to decrease over the winter season. Overall, the 2.5 years of cultivation caused a minor loss of soil Corg.
For Ntotal, crop cultivation of both winter rye and maize resulted in a decrease during the growing season, but this loss was restored over winter (p < 0.05). Crop diversification showed no significant effect on soil Ntotal (p > 0.05). Mineral N fertilization affected the Ntotal in 2019A and 2020S (p < 0.05), but not at other sampling dates. Ntotal reached a peak in 150N in 2019A and for the 250N treatment in 2020S, regardless of crop diversification.
The total soil C:N ratio was neither affected by N fertilization nor crop diversification (p > 0.05), except in the samples taken in 2019A, i.e. after cultivation of winter rye. Overall, the C:N ratio increased during crop growth and declined over winter (p < 0.05). Due to the split block design of the non-diversified and diversified cropping systems, the block effect was also considered, and there were no significant effects on physicochemical soil parameters detectable, with the one exception of Ntotal in the samples taken in 2020A, i.e. after maize with or without undersown ryegrass.
Effect of the N fertilization and crop diversification on soil microbial abundance
The mineral N fertilization affected fungal abundances consistently across all sampling dates (p < 0.05; Fig. 2; Table S1-2), while the effects on the abundance of bacteria and archaea were inconsistent. Crop diversification surprisingly showed a stronger influence on archaeal abundance than on bacteria and fungi. At three of the five sampling events, the archaeal gene copy numbers were different between the control and diversified cropping systems except for 2020S and 2020A (p < 0.05). In contrast, crop diversification affected the bacterial and fungal abundance only in 2019S (p < 0.05). The increased abundance of all three microbial groups in 2019S can be linked to a better microbial survival during winter in the presence of oil radish in the diversified system as compared to bulk soil in the non-diversified control. For bacterial abundance, significant differences were seen in only 3 of 15 cases (p < 0.05; Fig. 2), but interestingly these always indicated a positive effect by crop diversification; for fungi, five cases were significantly different, and four of them also indicated positive effects. In contrast, the archaea were the most responsive group to diversification in terms of abundance, and showed a negative response to diversification in seven out of eight cases.
Effect of inorganic N fertilization and crop diversification on the abundances of bacteria, archaea and fungi at the experimental field site, as assessed by qPCR from directly extracted soil DNA. Capital letters and lower-case letters indicate significant differences among different N fertilizations (T1: 0 kg N/ha; T4: 150 kg N/ha; T6: 250 kg N/ha; n = 4) and between two cropping systems (non-diversified control vs. crop diversification)
Overall, the abundance of soil bacteria, archaea and fungi appeared to be more strongly influenced by crop species and season than by N fertilization and crop diversification, respectively. During the winter rye season, the soil bacterial, archaeal and fungal abundances increased, but they decreased after the growth of maize (p < 0.05) (2020A). There was no significant block effect on soil microbial abundance.
The changes in bacterial (p < 0.05), archaeal (p < 0.001) and fungal (p < 0.01) abundances were highly associated with fluctuations of the pH, according to Spearman’s correlation coefficient. Positive correlations occurred in the range between pH 4.4 to 5.8. For Corg and Ntotal, there were positive correlations with bacterial (p < 0.05) and fungal (p < 0.05) abundance, but not archaeal abundances (p > 0.05). Furthermore, there was a negative correlation between abundances of both archaea and fungi, with the C:N ratio (p < 0.05).
Microbial carbon use efficiency
Neither mineral N fertilization nor crop diversification affected the soil microbial CUE in any of the three sampling events (2018A, 2019S, 2019A) analyzed (p > 0.05; Fig. 3; Table S3-4). However, crop diversification reduced microbial respiration and growth in the 2019A, and this was most pronounced with the 250N supply. At the beginning of the winter rye growing season (2019S), the N fertilization reduced microbial respiration, but only in the diversified cropping system (p < 0.05). After the winter rye growing season (2019A), N fertilization promoted microbial growth in the non-diversified cropping system (p < 0.05). This promotion was stronger with 250N than with 150N. Notably, CUE varied with crop species and season.
Effect of N fertilizations and crop diversification on soil microbial carbon use efficiency (CUE), microbial respiration and growth. Capital letters and lower-case letters indicate significant difference among different N fertilizations (T1: 0 kg N/ha; T4: 150 kg N/ha; T6: 250 kg N/ha; n = 4) and between two cropping systems (non-diversified control vs. crop diversification) according to LSD test, respectively. Three values are missing in microbial respiration and four in microbial growth due to technical problems
Across the three sampling events, microbial CUE was found to be driven by alternations of microbial growth rather than the changes of microbial respiration rates (Fig. 4). Strong negative correlations were found between microbial CUE value and soil Ntotal (p < 0.01), while the C:N ratio correlated positively with the CUE values (p < 0.05). In addition to the correlation with soil chemical parameters, the abundance of bacteria but not of archaea and fungi correlated positively with microbial CUE, respiration and growth (p < 0.001), respectively.
Spearman’s rank correlation matrix of physicochemical soil parameters, parameters of soil microbial metabolism and the relative abundance of microbial functional groups. Strong correlations are indicated by large circles; weak correlations are indicated by smaller circles. Positive correlations are displayed in blue and negative correlations in red. Both circle size and colour intensity are proportional to the spearman correlation coefficients. The correlation analysis was based on 72 replicates (three N treatment with four replicates in two cropping systems and all three sampling events combined)
Composition and diversity of the prokaryotic communities
In total, 3.63 million high-quality 16S rRNA gene amplicon sequences were obtained from the 96 samples included in this study. These grouped into 31,733 ASV. Sequences assigned to bacteria and archaea were 96.2% and 3.8%, respectively (Fig. S1). The most abundant bacterial phyla were Proteobacteria (relative abundance 29.2%), Actinobacteriota (21.8%), Acidobacteriota (11.8%), Bacteroidota (10.4%), Firmicutes (7.05%), Chloroflexi (5.19%), Verrucomicrobiota (4.50%), Planctomycetota (3.47%), Gemmatimonadota (3.12%), Myxococcota (2.34%) and Patescibacteria (1.12%). The dominant archaea were Crenarchaeota (99.0%).
Correlation analyses were conducted to evaluate the response of specific bacterial phyla to the different treatments and field conditions. Bacteroidota showed a strong positive correlation with C:N ratio (p < 0.001; Table S5). Furthermore, the relative abundance of Planctomycetota correlated positively with microbial respiration (p < 0.01) and microbial growth (p < 0.05), respectively. In contrast, the relative abundance of Firmicutes correlated negatively with microbial growth and CUE (p < 0.05). Myxococcota correlated negatively with soil pH (p < 0.01), and positively with microbial CUE (p < 0.05), microbial respiration and microbial growth (p < 0.01). In addition, the relative abundance of both Proteobacteria and Bacteroidota correlated negatively with microbial respiration (p < 0.05). On the other hand, microbial respiration correlated positively with the relative abundance of Gemmatimonadota (p < 0.05). Overall, the phylum-specific correlations suggest different adaptations of the soil microbiome to changing soil conditions as represented by the selected parameters.
Neither the level of N fertilization nor the crop diversification changed the community evenness (p > 0.05; Fig. S2). For the non-diversified cropping system, N fertilizations had no effect on the ASV richness and Shannon diversity, the latter indicated by the Shannon index (p > 0.05). In contrast, crop diversification increased both richness and Shannon diversity in the 150N treatment by the end of crop growing seasons (p < 0.05).
Effect of the N fertilization and crop diversification on the prokaryotic community and network structure
Non-metric multidimensional scaling (NMDS) plots were constructed to compare the overall differences in soil prokaryotic communities between different treatments (Fig. 5; Table S6). ANOSIM revealed that the community structures differed (p < 0.001) among different sampling events accompanied by distinct crop species and growth stages. The mineral N fertilization showed a significant impact on the prokaryotic community (p < 0.05) with the one exception of samples taken in spring 2020. Crop diversification significantly changed the community structure at the end of the growing season (p < 0.05) but not at the beginning (p > 0.05).
The identification of responsive prokaryotic taxa was performed at the family and ASV level (Fig. S3 and S4), but by means of pairwise comparison among three mineral N treatments and two cropping systems in four sampling events, no significantly responding ASV or family were detected.
Correlation networks of co-occurring ASV were used to explore assemblage patterns under different conditions of N fertilization and crops (Fig. 6 and Fig. S5; Table 2). The network constructed with data from all four sampling times revealed a striking difference between the non-diversified and the diversified cropping systems at the conventional 150N supply: Networks of the diversified system were characterized by a higher number of nodes, edges, and other descriptors demonstrating a much more elaborated structure.
Co-occurrence networks of amplicon sequence variants (ASV) representing the prokaryotic communities across four sampling events in different treatments based on average ASV numbers of above 1 in all samples. Connections indicate strong (r > 0.7 or r < − 0.7) and significant (corrected p-value < 0.05) correlations. The positive and negative correlations between nodes are displayed with red and blue lines, respectively. Networks were constructed based on 16 replicates (each treatment, with four replicates and all four sampling events combined). For network variants based on higher average ASV see Supplemental material (Fig. S5)
Effect of the N fertilization and crop diversification on suspected microbial functional groups
FAPROTAX analysis allowed to assign 17.8% (5,640 out of 31,733) of all ASV to at least one potential ecological type. Overall, those assigned ASV encompassed a total of 63 different ecological types, with chemo-heterotrophy and aerobic chemo-heterotrophy being the dominant functions. A total of 14 ecological types which represented functional groups mainly contributing to the C and N cycles were selected for further analyses, as these functions were considered as being most relevant for the arable land use in this study (Fig. 4 and Fig. S6). Except for nitrification, N-transforming functional groups involved in nitrogen respiration, denitrification, nitrate reduction, nitrate respiration and nitrite respiration, responded in a similar way. Crop diversification showed strong effects on increasing the relative abundance of taxa associated with nitrification, while decreasing the proportion of nitrogen and nitrate respiration, nitrate reduction and aromatic compound degradation functional groups in the excessive 250N treatment. This effect was most pronounced with samples from 2019A, i.e., after winter rye with or without undersown ryegrass. The relative abundance of ASV associated with N fixation correlated negatively with soil pH (p < 0.05) but positively with Corg (p < 0.05) and Ntotal (p < 0.05). ASV associated with methanol oxidation and aromatic compound degradation correlated positively with the microbial CUE, growth and respiration, while the ureolysis showed a negative relationship with those parameters.
Discussion
The cultivation of crops on arable land has immediate consequences for the living conditions of the soil microbiome. Soil tillage, the addition of fertilizers and the sequences of different crops with their particular root architectures and organic exudations all together can change soil microhabitats and, thus, are likely of inducing adaptation processes emerging in structurally and functionally different microbiomes. Both soil pH and organic nutrients have been shown to act as major selective factors for shaping the soil microbiome (Fierer and Jackson 2006; Szoboszlay et al. 2017). In our study, along with a sequence of cultivating potato, winter rye and maize, we observed shifts of the soil pH in a range between 4.4 and 5.8. The application of mineral N fertilizer caused soil acidification, which is also known from other studies (Schroder et al. 2011; Snapp and Surapur 2018; Yang et al. 2018). Thus, as a consequence of mineral fertilization, the microbiome does not only receive an additional source of N, but also faces a decrease in pH. While additional N will change the soil C:N ratio, which could promote growth of N-limited microorganisms, the decline in pH may also result in mobilization of otherwise soil-adsorbed nutrients, due to weakening anionic bonds with clay minerals by a decline of bivalent cations i.e. Ca2+ or Mg2+ (Goulding 2016). In fact, a positive growth effect of additional N was seen for fungal abundance, but not for bacteria or archaea. To merely explain this effect by the additional N would be surprising, considering that fungi have a higher C:N ratio (Six et al. 2006) and therefore lower N requirement than bacteria (Holland and Coleman 1987). But it was already demonstrated that the availability of N would not always suffice to explain differential growth between fungi and bacteria (Rousk and Baath 2007). Possibly, the decrease in pH mobilized C adsorbed at mineral surfaces, which would preferentially be metabolized by fungi and thus support their growth rather than growth of bacteria.
A lack of consistent N fertilizer impact on bacterial growth was also indicated by the fact that there were no clear N-induced compositional shifts in the prokaryotic community structure (which was represented to 96% by bacteria). Furthermore, neither microbial growth, as measured by incorporation of 18O from H218O into DNA, nor microbial respiration or the CUE were significantly and continually affected by the N additions. Apparently, these results contradicted the conclusion on N-stimulated fungal growth obtained with qPCR. The different techniques and their respective sensitivities used in this study remain to be determined, for characterizing properties of the soil microbiome under different environmental conditions. Even the compositional analyses of the prokaryotic microbiome determined in this study by massively parallel 16S rRNA gene amplicon sequencing failed to detect tangible effects of the N fertilizations on the community structure, thus supporting results obtained with respiration, microbial growth and CUE. Especially considering the two-step PCR for sequencing preparation was used in this study, the response of low-abundant microorganisms to N fertilization and crop diversification might be underestimated (Finn et al. 2022).
Possibly the failure to detect a fertilizer effect on the prokaryotic community structure was influenced by the fact that we sampled bulk soil rather than rhizosphere, since we wanted to consider the impact on the whole soil community and not only on a specific compartment. The sampling of bulk soil rather than rhizosphere in this study probably captured more 16S rRNA gene signals from inactive or even dead bacterial cells, thus masking the indirect effects triggered by root exudation. Furthermore, it should be noted that the experimental design of this agricultural field study only allowed four replicates for each treatment, which probably limited our capacity to detect less pronounced but significant effects, given the typical intrinsic environmental variation under field conditions.
The cropping system analyzed in this study included a regionally common rotation with potato, winter rye and maize. Comparisons between spring and autumn samples indicated that the cultivation of winter rye increased soil pH while maize decreased it. Compared to the non-diversified control, the diversification included undersown ryegrass in both winter rye and maize season, and in addition during the winter season, oilseed radish. The diversification was suspected to have a positive effect by translocation of more photosynthetically fixed organic C into soil due to higher root density and rhizodeposition. This striking effect was clearly seen in this study after the winter season in which the diversified system, with its oilseed radish, supported a relatively higher gene copy numbers than in the non-diversified control, thereby indicating a higher microbial population during winter. Since the soil microbiome can also provide nutrients, e.g. in the subsequent growing season, the maintenance of a higher biomass during winter should be ecologically and economically beneficial. Similar beneficial effects of the over-winter stabilization of the soil microbiome under diversified systems were observed in a long-term field study, where in addition to cover crops, crop residues were returned to soil (Liu et al. 2022).
With cultivation of winter rye, we observed an increase in soil Corg while the cultivation of maize caused a decline. Such variation can be explained by the specific root architectures of the respective crops, their capacity to assimilate soil nutrients and specific agricultural management practices, including time of fertilization and soil tillage. Interestingly, the shifts of Corg in both directions were less pronounced with the diversified cropping system with its undersown ryegrass. These additional plants apparently increased the higher buffering capacity in soils, resulting in less variable soil microhabitat conditions. Due to the stabilized microhabitat conditions along with additional rhizodepositions of the ryegrass we expected that diversified systems support the presence of more complex and less changeable microbiomes.
In the non-diversified system, N fertilization had no effect on the prokaryotic richness and diversity, but in the diversified systems both parameters increased. This beneficial effect can be explained by the additional rhizodepositions which provide most likely other C sources than the main crops, and generating more diverse soil microhabitats, thus promoting additional bacterial taxa (ASV) to grow as compared to the non-diversified system. However, despite the clear indication by the diversity and richness indices, we were unable in this study to identify specific taxa or phylogenetic groups specifically promoted by the diversified systems. A statistically robust identification of indicator taxa probably would have required a higher number of replicates and/or annual replications, considering that field studies have a higher variation than simple systems, i.e. microcosms.
While we could not unravel soil bacterial indicators for diversification, ASV association networks provided a clear indication that the prokaryotic communities were more elaborated and complex after addition of conventional amounts of N fertilizer in soils of diversified cropping systems than in non-diversified controls. This difference can be explained by growth of a more diverse prokaryotic community which takes advantage of a higher diversity of rhizodepositions in soils of the diversified cropping system. With the excessive N-supply such benefits of diversified cropping systems on the prokaryotic association networks, however, disappeared, perhaps because higher N supplies would activate the decomposition of additional organic C from soil. Interestingly, at this excessive N supply, the functional prediction model that we used in this study, i.e. the FAPROTAX tool, indicated at one sampling event, i.e. after cultivation of rye, strong differences caused by the diversification (undersown ryegrass), a shift in the N cycle, by increasing the relative abundance of nitrifiers in comparison to denitrifiers. Although this predictive tool certainly cannot replace whole metagenome analyses, this model was useful in other studies to estimate microbial metabolic phenotypes and it showed a relatively good consistency with metagenomics results (Louca et al. 2016; Sansupa et al. 2021; Wei et al. 2020). While it would be too speculative to explain a potential promotion of nitrifiers by the diversified cropping system in this study, the data indicate that most likely functional changes in the microbiome occur when non-diversified cropping systems are replaced by diversified.
In contrast to the indicated changes of the prokaryotic community in response to crop diversification, CUE, as determined in this study, was not affected. The values varied between crops and seasons, but not with diversification. This is in accordance to conclusions obtained analyzing factors driving the abundance of bacteria, archaea and fungi. The strong correlations seen between bacterial abundance and CUE suggest that the value was mainly driven by bacteria, even though fungi are also known to strongly contribute to respiration, i.e. one of the major factors determining the CUE. At the end of one growing season (2019A), crop diversification decreased respiration, most pronounced with the highest N supply, but this effect was not tangible at other sampling occasions. Thus, we could not confirm in this study that diversification promoted CUE and thereby the stabilization of a microbiome in soil. Considering that the diversification treatments at the field site were started only three years before the onset of this study, the time period for selecting characteristic soil microbiomes may not have been long enough.
Conclusion
The diversified cropping system showed positive effects on the prokaryotic soil microbiome at conventional N application rates, specifically as a higher richness, diversity, and a more elaborated network structure, rather than by promoting growth of specific indicator taxa. The positive effects of the diversified systems can be attributed to the higher buffering of soil pH and a suspected higher amount and diversity of exudates due to the additional soil input of photosynthetically fixed organic C by higher root density and rhizodeposition. Thereby the results of this study underline the beneficial effects of crop diversification on the soil microbiomes and their ecosystem.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Beaudoin N, Saad JK, Van Laethem C, Machet JM, Maucorps J, Mary B (2005) Nitrate leaching in intensive agriculture in Northern France: Effect of farming practices, soils and crop rotations. Agr Ecosyst Environ 111:292–310. https://doi.org/10.1016/j.agee.2005.06.006
Bicharanloo B, Shirvan MB, Keitel C, Dijkstra FA (2020) Rhizodeposition mediates the effect of nitrogen and phosphorous availability on microbial carbon use efficiency and turnover rate. Soil Biol Biochem 142. https://doi.org/10.1016/j.soilbio.2020.107705
Bolyen E, Rideout JR, Dillon MR, Bokulich N, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo JR, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang LJ, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, vander Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan YH, Wang MX, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang YL, Zhu QY, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581. https://doi.org/10.1038/Nmeth.3869
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci USA 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
Csardi G, Nepusz T (2006) The igraph software package for complex network research. Int J Complex Syst 1695:1–9. http://igraph.org
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK (2016) Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun 7. https://doi.org/10.1038/ncomms10541
Denef K, Roobroeck D, Wadu MCWM, Lootens P, Boeckx P (2009) Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol Biochem 41:144–153. https://doi.org/10.1016/j.soilbio.2008.10.008
Erisman JW, Sutton MA, Galloway J, Klimont Z, Winiwarter W (2008) How a century of ammonia synthesis changed the world. Nat Geosci 1:636–639. https://doi.org/10.1038/ngeo325
Fernandes AD, Reid JNS, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB (2014) Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. https://doi.org/10.1186/2049-2618-2-15
Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB (2013) ANOVA-Like Differential Gene Expression Analysis of Single-Organism and Meta-RNA-Seq. Plos One 8. https://doi.org/10.1371/2/journal.pone.0067019
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. P Natl Acad Sci USA 103:626–631. https://doi.org/10.1073/pnas.0507535103
Finn DR, Samad MS, Tebbe CC (2022) One-step PCR amplicon sequencing libraries perform better than two-step when assessing soil microbial diversity and community profiles. Fems Microbiol Lett 369. https://doi.org/10.1093/femsle/fnac079
Foyer CH, Lam HM, Nguyen HT, Siddique KHM, Varshney RK, Colmer TD, Cowling W, Bramley H, Mori TA, Hodgson JM, Cooper JW, Miller AJ, Kunert K, Vorster J, Cullis C, Ozga JA, Wahlqvist ML, Liang Y, Shou HX, Shi K, Yu JQ, Fodor N, Kaiser BN, Wong FL, Valliyodan B, Considine MJ (2016) Neglecting legumes has compromised human health and sustainable food production. Nat Plants 2. https://doi.org/10.1038/Nplants.2016.112
Francioli D, Schulz E, Lentendu G, Wubet T, Buscot F, Reitz T (2016) Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front Microbiol 7. https://doi.org/10.3389/fmicb.2016.01446
Geyer KM, Dijkstra P, Sinsabaugh R, Frey SD (2019) Clarifying the interpretation of carbon use efficiency in soil through methods comparison. Soil Biol Biochem 128:79–88. https://doi.org/10.1016/j.soilbio.2018.09.036
Goulding KWT (2016) Soil acidification and the importance of liming agricultural soils with particular reference to the United Kingdom. Soil Use Manage 32:390–399. https://doi.org/10.1111/sum.12270
Grenié M, Denelle P, Tucker CM, Munoz F, Violle C (2017) funrar: An R package to characterize functional rarity. Divers Distrib 23:1365–1371. https://doi.org/10.1111/ddi.12629
Hall MD, Shaffer MJ, Waskom RM, Delgado JA (2001) Regional nitrate leaching variability: What makes a difference in northeastern Colorado. J Am Water Resour Assoc 37:139–150. https://doi.org/10.1111/j.1752-1688.2001.tb05481.x
Hartmann M, Six J (2022) Soil structure and microbiome functions in agroecosystems. Nat Rev Earth Env. https://doi.org/10.1038/s43017-022-00366-w
Hauggaard-Nielsen H, Mundus S, Jensen ES (2012) Grass-clover undersowing affects nitrogen dynamics in a grain legume cereal arable cropping system. Field Crop Res 136:23–31. https://doi.org/10.1016/j.fcr.2012.07.001
He TX, Xie DT, Ni JP, Li Z, Li ZL (2020) Nitrous oxide produced directly from ammonium, nitrate and nitrite during nitrification and denitrification. J Hazard Mater 388. https://doi.org/10.1016/j.jhazmat.2020.122114
Herbold C, Pelikan C, Kuzyk O, Hausmann B, Angel R, Berry D, Loy A (2015) A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front Microbiol 6. https://doi.org/10.3389/fmicb.2015.00731
Holland EA, Coleman DC (1987) Litter Placement Effects on Microbial and Organic-Matter Dynamics in an Agroecosystem. Ecology 68:425–433. https://doi.org/10.2307/1939274
Joergensen RG (1996) The fumigation-extraction method to estimate soil microbial biomass: Calibration of the k(EC) value. Soil Biol Biochem 28:25–31. https://doi.org/10.1016/0038-0717(95)00102-6
Kankanen H, Eriksson C (2007) Effects of undersown crops on soil mineral N and grain yield of spring barley. Eur J Agron 27:25–34. https://doi.org/10.1016/j.eja.2007.01.010
Kim N, Zabaloy MC, Guan KY, Villamil MB (2020) Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol Biochem 142. https://doi.org/10.1016/j.soilbio.2019.107701
Kuypers MMM, Marchant HK, Kartal B (2018) The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. https://doi.org/10.1038/nrmicro.2018.9
Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmerTest Package: Tests in Linear Mixed Effects Models. J Stat Softw 82:1–26. https://doi.org/10.18637/jss.v082.i13
Liu B, Arlotti D, Huyghebaert B, Tebbe CC (2022) Disentangling the impact of contrasting agricultural management practices on soil microbial communities – Importance of rare bacterial community members. Soil Biol Biochem 166. https://doi.org/10.1016/j.soilbio.2022.108573
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277. https://doi.org/10.1126/science.aaf4507
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetj 17:10–12. https://doi.org/10.14806/ej.17.1.200
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. Bmc Microbiol 5. https://doi.org/10.1186/1471-2180-5-28
Mendiburu F (2021) agricolae: Statistical Procedures for Agricultural Research. R package version 1.3–5. https://cran.r-project.org/web/packages/agricolae/index.html
Momesso L, Crusciol CAC, Cantarella H, Tanaka KS, Kowalchuk GA, Kuramae EE (2022) Optimizing cover crop and fertilizer timing for high maize yield and nitrogen cycle control. Geoderma 405. https://doi.org/10.1016/j.geoderma.2021.115423
Muhammad I, Wang J, Sainju UM, Zhang SH, Zhao FZ, Khan A (2021) Cover cropping enhances soil microbial biomass and affects microbial community structure: A meta-analysis. Geoderma 381. https://doi.org/10.1016/j.geoderma.2020.114696
Poeplau C, Helfrich M, Dechow R, Szoboszlay M, Tebbe CC, Don A, Greiner B, Zopf D, Thumm U, Korevaar H, Geerts R (2019) Increased microbial anabolism contributes to soil carbon sequestration by mineral fertilization in temperate grasslands. Soil Biol Biochem 130:167–176. https://doi.org/10.1016/j.soilbio.2018.12.019
Qi JY, Jensen JL, Christensen BT, Munkholm LJ (2022) Soil structural stability following decades of straw incorporation and use of ryegrass cover crops. Geoderma 406. https://doi.org/10.1016/j.geoderma.2021.115463
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Revelle W (2022) psych: Procedures for Psychological, Psychometric, and Personality Research. Northwestern University, Evanston, Illinois
Rocha KF, Kuramae EE, Borges BMF, Leite MFA, Rosolem CA (2020) Microbial N-cycling gene abundance is affected by cover crop specie and development stage in an integrated cropping system. Arch Microbiol 202:2005–2012. https://doi.org/10.1007/s00203-020-01910-2
Rognes T, Flouri T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. Peerj 4. https://doi.org/10.7717/peerj.2584
Rousk J, Baath E (2007) Fungal and bacterial growth in soil with plant materials of different C/N ratios. Fems Microbiol Ecol 62:258–267. https://doi.org/10.1111/j.1574-6941.2007.00398.x
Sansupa C, Wahdan SFM, Hossen S, Disayathanoowat T, Wubet T, Purahong W (2021) Can We Use Functional Annotation of Prokaryotic Taxa (FAPROTAX) to Assign the Ecological Functions of Soil Bacteria?. Appl Sci-Basel 11. https://doi.org/10.3390/app11020688
Schroder JL, Zhang HL, Girma K, Raun WR, Penn CJ, Payton ME (2011) Soil Acidification from Long-Term Use of Nitrogen Fertilizers on Winter Wheat. Soil Sci Soc Am J 75:957–964. https://doi.org/10.2136/sssaj2010.0187
Schroeder J, Kammann L, Helfrich M, Tebbe CC, Poeplau C (2021) Impact of common sample pre-treatments on key soil microbial properties. Soil Biol Biochem 160. https://doi.org/10.1016/j.soilbio.2021.108321
Simon E, Canarini A, Martin V, Seneca J, Bockle T, Reinthaler D, Potsch EM, Piepho HP, Bahn M, Wanek W, Richter A (2020) Microbial growth and carbon use efficiency show seasonal responses in a multifactorial climate change experiment. Commun Biol 3. https://doi.org/10.1038/s42003-020-01317-1
Six J, Frey SD, Thiet RK, Batten KM (2006) Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci Soc Am J 70:555–569. https://doi.org/10.2136/sssaj2004.0347
Smith CM, David MB, Mitchell CA, Masters MD, Anderson-Teixeira KJ, Bernacchi CJ, DeLucia EH (2013) Reduced Nitrogen Losses after Conversion of Row Crop Agriculture to Perennial Biofuel Crops. J Environ Qual 42:219–228. https://doi.org/10.2134/jeq2012.0210
Snapp S, Surapur S (2018) Rye cover crop retains nitrogen and doesn’t reduce corn yields. Soil till Res 180:107–115. https://doi.org/10.1016/j.still.2018.02.018
Spohn M, Potsch EM, Eichorst SA, Woebken D, Wanek W, Richter A (2016) Soil microbial carbon use efficiency and biomass turnover in a long-term fertilization experiment in a temperate grassland. Soil Biol Biochem 97:168–175. https://doi.org/10.1016/j.soilbio.2016.03.008
Szoboszlay M, Dohrmann AB, Poeplau C, Don A, Tebbe CC (2017) Impact of land-use change and soil organic carbon quality on microbial diversity in soils across Europe. Fems Microbiol Ecol 93. https://doi.org/10.1093/femsec/fix146
Thorup-Kristensen K, Dresboll DB, Kristensen HL (2012) Crop yield, root growth, and nutrient dynamics in a conventional and three organic cropping systems with different levels of external inputs and N re-cycling through fertility building crops. Eur J Agron 37:66–82. https://doi.org/10.1016/j.eja.2011.11.004
Tonitto C, David MB, Drinkwater LE (2006) Replacing bare fallows with cover crops in fertilizer-intensive cropping systems: A meta-analysis of crop yield and N dynamics. Agr Ecosyst Environ 112:58–72. https://doi.org/10.1016/j.agee.2005.07.003
Toth JD, Fox RH (1998) Nitrate losses from a corn-alfalfa rotation: Lysimeter measurement of nitrate leaching. J Environ Qual 27:1027–1033. https://doi.org/10.2134/jeq1998.00472425002700050007x
Valkama E, Lemola R, Kankanen H, Turtola E (2015) Meta-analysis of the effects of undersown catch crops on nitrogen leaching loss and grain yields in the Nordic countries. Agr Ecosyst Environ 203:93–101. https://doi.org/10.1016/j.agee.2015.01.023
Vance ED, Brookes PC, Jenkinson DS (1987) An Extraction Method for Measuring Soil Microbial Biomass-C. Soil Biol Biochem 19:703–707. https://doi.org/10.1016/0038-0717(87)90052-6
Vitousek PM, Hattenschwiler S, Olander L, Allison S (2002) Nitrogen and nature. Ambio 31:97–101. https://doi.org/10.1579/0044-7447-31.2.97
Wei GS, Li MC, Shi WC, Tian RM, Chang CY, Wang ZR, Wang NX, Zhao GX, Gao Z (2020) Similar drivers but different effects lead to distinct ecological patterns of soil bacterial and archaeal communities. Soil Biol Biochem 144. https://doi.org/10.1016/j.soilbio.2020.107759
Wickham H, François R, Henry L, Müller K (2021) dplyr: A Grammar of Data Manipulation. R package version 1.0.7. https://cran.r-project.org/web/packages/dplyr/index.html
Widdig M, Schleuss PM, Biederman LA, Borer ET, Crawley MJ, Kirkman KP, Seabloom EW, Wragg PD, Spohn M (2020) Microbial carbon use efficiency in grassland soils subjected to nitrogen and phosphorus additions. Soil Biol Biochem 146. https://doi.org/10.1016/j.soilbio.2020.107815
Yang XD, Ni K, Shi YZ, Yi XY, Zhang QF, Fang L, Ma LF, Ruan JY (2018) Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agr Ecosyst Environ 252:74–82. https://doi.org/10.1016/j.agee.2017.10.004
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO (2014) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. https://doi.org/10.1093/nar/gkt1209
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. https://doi.org/10.1002/bit.20347
Acknowledgements
Jana Usarek, Britta Müller and Karin Trescher supported this work with their excellent technical support, which we gratefully acknowledge. We would also like to thank the technical staff of the Thünen Institute involved in the determination of soil microbial carbon use efficiency: Sabine Wathsack and Kerstin Gilke. Thanks to Jens Dyckmans and his colleagues at the Centre for Stable Isotope Research and Analysis at the University of Göttingen for supporting this work. We thank the Chamber of Agriculture Lower Saxony for giving us access to their excellently maintained field experiment in Hamerstorf, Germany and generously supporting our visits during sampling events.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by EU Horizon 2020 project DiverIMPACTS—Diversification through Rotation, Intercropping, Multiple Cropping, Promoted with Actors and value-Chains towards Sustainability (Grant agreement No 727482).
Author information
Authors and Affiliations
Contributions
Christoph C. Tebbe conceptualized the experimental question and defined the objectives and analytical tools. Hauke Ahnemann carried out the field experiments. Soil sampling, lab work, data collection and analysis were performed by Bei Liu. Julia Schroeder and Christopher Poeplau guided the work of soil microbial carbon use efficiency determination. Christoph C. Tebbe supervised the project. The first draft of the manuscript was written by Bei Liu and Christoph C Tebbe, and all authors commented on previous versions of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Tida Ge.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, B., Schroeder, J., Ahnemann, H. et al. Crop diversification improves the diversity and network structure of the prokaryotic soil microbiome at conventional nitrogen fertilization. Plant Soil 489, 259–276 (2023). https://doi.org/10.1007/s11104-023-06011-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06011-9