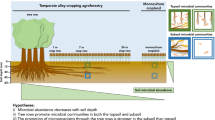
Abstract
Aims
Cropland agroforestry systems are land-use systems with numerous environmental advantages over monoculture croplands including promotion of soil life. This study aimed to investigate tree-species and tree-distance effects on soil biota in a temperate agroforestry system.
Methods
Our study was conducted at a paired alley-cropping and monoculture cropland system. The tree rows of the agroforestry system comprised of blocks of poplar Fritzi Pauley, poplar Max 1 or black locust. Within the agroforestry system, soil microbial and earthworm communities were collected along transects spanning from the center of the tree rows into the crop rows. Archaea, bacteria, and fungi were quantified using real-time PCR. The community composition of fungi and earthworms was deciphered using amplicon sequencing and morphological identification, respectively.
Results
Tree rows promoted the abundance of bacteria and earthworms, which we attribute mainly to tree litter input and the absence of tillage. Fungal community composition was altered by the tree rows, resulting in an increased proportion of ectomycorrhizal fungi in the tree-row associated mycobiome. The proportion of Blumeria graminis, the causal agent of powdery mildew, increased with increasing distance from the trees. We suggest that enhanced microbial antagonism, increased earthworm densities and/or altered microclimate contributed to the suppression of B. graminis in vicinity of the trees. Tree-species effect had a minor influence on the abundance and composition of soil communities at our study site.
Conclusions
In comparison to monoculture cropland, agroforestry benefits the abundance, diversity, and function of soil biota and may enhance soil suppressiveness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agroforestry systems are land-use systems that combine trees with crops and/or livestock. In the temperate zone, alley-cropping agroforestry systems that alternate rows of trees with rows of crops are gaining popularity. Tree rows of these systems usually consist of either fast-growing trees (e.g. poplar or willow) for biomass production or quality hardwoods such as cherry or walnut trees. The environmental benefits of temperate agroforestry practice are well recognized and include, inter alia, increased carbon sequestration (e.g. Mayer et al. 2022), reduced nutrient leaching (e.g. Allen et al. 2004; Wang et al. 2011), reduced soil erosion, and promotion of biodiversity (e.g. Varah et al. 2013, 2020). Certain advantages of alley-cropping systems over monoculture croplands, such as the complementary use of resources, are due to interspecific interactions between the trees and the crops (Jose et al. 2004). For example, deep-rooting trees are able to take up nutrients leached below the rooting zone of the crops (‘safety-net’-function of the trees; Allen et al. 2004) and make them available to crops via litter fall. This process was recently dubbed as ‘nutrient pumping’ (Isaac and Borden 2019) and is supported by findings of increased soil nutrient availability in vicinity of the tree rows of temperate agroforestry systems (Pardon et al. 2017).
Soil biota are known to predominantly benefit from temperate agroforestry practice (Beule et al. 2022a; Marsden et al. 2020). Several studies on soil microorganisms in alley-cropping systems revealed that the tree rows promote microbial population size and that this effect is not only limited to the tree rows but extends gradually into the crop rows (i.e. stronger promotion in vicinity of the trees) (e.g. Beule et al. 2022b; Guillot et al. 2021). Furthermore, soil microbiome studies reported that the composition of bacterial and fungal communities strongly differs between the tree and crop rows (e.g. Beule et al. 2021; Beule & Karlovsky 2021a). Based on these studies, it was recently reviewed that alley-cropping systems increase microbial diversity in soil through increased beta diversity rather than alpha diversity (Beule et al. 2022a). Recent articles showed that soil fungal communities strongly respond to temperate agroforestry practices. For example, one study found that Basidiomycota in soil were up to 330 times more abundant in poplar tree rows of alley-cropping systems as compared to adjacent cropland monocultures (Beule et al. 2021). The same study used amplicon sequencing to investigate the composition of the soil fungal community and found that certain ectomycorrhizal fungi (Cortinarius, Geopora, and Inocybe) were particularly promoted by poplar trees, which holds great potential to improve nutrient acquisition in agroforestry systems (Beule et al. 2021). In the present study, we aimed to explore the impact of different tree species on soil fungal communities in agroforestry systems.
In 1999, Seiter and co-workers (1999) observed that most of the tree leaf litter in an alder–maize alley-cropping system was pulled into the burrows by Lumbricus terrestris but did not provide data to substantiate their field observation. Few months earlier, Price and Gordon (1998) published an article demonstrating tree-species specific promotion of earthworms through tree rows in an alley-cropping system. Since then, greater densities of earthworms in tree rows of temperate agroforestry systems as compared to bordering crop rows or adjacent monoculture croplands have been reported (Cardinael et al. 2019; D'Hervilly et al. 2022). Yet, studies investigating earthworm communities in temperate agroforestry systems along fine spatial gradients from the tree rows into the crop rows comprising more than two distances from the tree rows are scarce. Furthermore, most temperate agroforestry systems feature either a single tree species or an intermixture of tree species (e.g. Cardinael et al. 2019), thus disabling direct comparisons among tree species within the same system. To our best knowledge, the investigation by Price and Gordon (1998) is the only study to assess earthworm communities under different tree species within the same temperate agroforestry system. However, the authors did not report species identities or ecological groups of collected earthworms. Recent studies indicated differences in the response of different ecological groups of earthworms to agroforestry practice (Cardinael et al. 2019; D'Hervilly et al. 2022), highlighting the importance of investigating them.
In the present study, we chose two poplar species, as poplar trees are the most commonly planted short-rotation trees in modern alley-cropping agroforestry systems in the temperate zone. Although not commonly planted in temperate agroforestry systems, we further chose black locust as it is a fast-growing, nitrogen-fixing tree species. We expect differences in overall soil biota communities under different tree species because root architecture, litter quality, and soil-nutrient cycling are tree species-specific (e.g. Das and Chaturvedi 2008, Aponte et al. 2013, Borden et al. 2017).
This study aimed to investigate the impact of three different tree species (two poplar species and black locust) on representatives of the soil biota community (archaea, bacteria, fungi, and earthworms) in a temperate alley-cropping agroforestry system. Furthermore, tree-distance effects on soil biota were tested by sampling multiple locations along transects spanning from the tree row into the crop row of the agroforestry system. We hypothesized that tree rows promote the abundance and alter the community composition of soil biota. We further hypothesized that these changes are dependent on the distance to the trees (tree-distance effect) as well as the tree species (tree-species effect).
Materials and Methods
Study site and sampling design
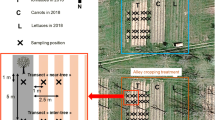
Our study site was located on a Gleyic Cambisol soil (IUSS Working Group WRB 2015) near Forst, Brandenburg, Germany (51°47′11″N, 14°38′05″E; m.a.m.s.l.: 67 m; mean annual temperature: 9.6 ± 0.2 °C; mean annual precipitation: 568 ± 21 mm; Fig. 1A), which is located in the glacially influenced region of the North German Plain. General biochemical and physical soil properties of the site were recently characterized by Schmidt et al. (2021). According to aerial images, the study site was under agricultural use for at least 50 years prior to conversion of cropland monoculture to alley-cropping agroforestry. At the study site, a conventionally managed alley-cropping agroforestry system was spatially paired with a conventional monoculture cropland system that served as a reference land use. The agroforestry system was established in 2010 and was 12 years old during sampling. The system consisted of 12 m-wide rows of trees (north–south orientation) that were alternated with 48 m-wide rows of arable crops (Fig. 1B). Tree rows were hand planted and consisted of three different tree species. The tree rows comprised four individual rows of trees (Fig. 1 C, D, E). The tree rows that defined the crop row consisted of three different tree species that were planted in alternating segments at a length of approx. 165 m (Fig. 1B). The different tree species were i) Populus trichocarpa cv. Fritzi Pauley (referred to as ‘poplar Fritzi Pauley’; Fig. 1C), ii) P. nigra × P. maximowiczii cv. Max 1 (referred to as ‘poplar Max 1’; Fig. 1D), and iii) black locust (Robinia pseudoacacia) (referred to as ‘black locust’; Fig. 1E). The aboveground biomass of the trees was harvested using a forage harvester in February 2015, March 2018, and February 2021. Tree harvests were conducted when the soil was frozen to avoid soil compaction by the machinery.
Study site and study design. Location of the study site near Forst (federal state of Brandenburg), Germany (A), study design (B), and photos of the three different tree species cultivated at the site (poplar Fritzi Pauley (C), poplar Max 1 (D), and black locust (E)) taken in April 2022. Photo credit: Lukas Beule
The management of the crop rows was identical to that of the monoculture cropland. The crop rotation was maize (Zea mays) – summer barley (Hordeum vulgare) – summer oat (Avena strigosa) – winter wheat (Triticum aestivum) – winter barley (H. vulgare) (2018 – 2019 – 2020 – 2021 – 2022). In March 2022, the crop row of the agroforestry system and the monoculture cropland received 40 – 0 – 0 kg N – P – K ha−1 in the form of mineral fertilizer. Maize and small-grain cereal crops were harvested with a standard 24-m wide combine harvester. Crop rows and monoculture cropland were conventionally ploughed at a depth of 25 cm.
It is well established that trees in alley-cropping systems introduce spatial heterogeneity (e.g. Guillot et al. 2021; Beule et al. 2020). Thus, samples in the agroforestry system were collected along four transects (replicate plots) per tree species spanning from the tree row into the crop row. Samples were collected in the center of the tree row as well as at 1 m, 7 m, and 18 m distance from the trees within the crop row (3 tree species × 4 transects × 4 sampling locations within each transect = 48 samples; Fig. 1B). In the adjacent monoculture cropland, samples were collected in the center of each replicate plot (4 samples; Fig. 1B).
Soil and earthworm sampling
Soil samples for the determination of the microbial community and general soil properties (soil pH, soil organic carbon (SOC), total N, extractable nutrients) in the upper 5-cm topsoil were collected on 23 February 2022 (prior to fertilization). Three soil samples with a volume of 250 cm3 were collected at each sampling location using stainless steel cylinders (5 cm height), transferred to a sterile polyethylene bag, and thoroughly homogenized. An aliquot of approx. 50 g fresh soil was transferred into a sterile 50-ml Falcon tube and frozen at -20 °C in the field. Upon arrival at the laboratory, samples were stored at -20 °C until freeze-drying for 48 h. The freeze-dried material was finely ground using a vortexer (Beule et al. 2019a) and stored at -20 °C until DNA extraction. The remaining fresh soil in the polyethylene bag was used for determination of the general soil properties. Since earthworm activity at our site is generally low in February, earthworm communities were sampled on 11 and 12 April 2022 as described below (see Extraction of earthworms). Soil samples (250 cm3) for the determination on soil bulk density and water-filled pore space (WFPS) were collected on all sampling dates and sampling locations (see above).
Extraction of earthworms
Earthworms were directly extracted from soil by applying 5 L of 0.01% (w/v) allyl isothiocyanate (AITC) solution on an area of a quarter square meter as described by Zaborski (2003). The extractant was prepared in the field by adding 500 mg AITC (pre-dissolved in 50 ml isopropanol (w/v)) to 4.95 L tap water immediately before extraction. To ensure that the extractant was applied only to a surface of 0.25 m2, it was poured into a 50 × 50 cm open metal frame which was embedded approx. 5 cm deep into the soil (Figure S1 A). To allow better monitoring of extracted worms, plant material within the frame was carefully removed prior to the application of AITC. Following the AITC application, the soil surface within the frame was continuously monitored for at least 30 min and any surfacing earthworms were collected using tweezers, thoroughly washed, and stored in tap water (Figure S1 B). Upon arrival to the laboratory, earthworms were stored at 5 °C in the dark until morphological identification. Morphological identification and recording of the fresh weight (including gut content) of each individual earthworm were carried out within 48 h post sampling and earthworms were subsequently released. Furthermore, earthworms were classified into three ecological groups: anecic, endogeic, and epigeic species, which were introduced by Bouché in 1972. Earthworm data have been deposited at the BonaRes repository (https://doi.org/10.20387/bonares-y3je-zz30).
Soil DNA extraction
DNA from soil was extracted using a CTAB-based protocol with phenol and chloroform/isoamyl alcohol extraction (Beule et al. 2019a). Briefly, 50 mg finely ground soil was suspended in a mixture containing 1 ml CTAB, 1 µL 2-mercaptoethanol, and 2 µL pronase E (30 mg/ml). The mixture was incubated at 42 °C and subsequently at 65 °C for 10 min each and 900 µL phenol were added. The mixture was shaken, centrifuged at 4,600 × g at room temperature for 10 min, and 800 µL of the supernatant were transferred into a new 2 mL tube. 800 µL chloroform/isoamyl alcohol (24:1 (v/v)) were added, the mixture was shaken, incubated on ice for 10 min, and centrifuged at 4,600 × g at room temperature for 10 min. Following centrifugation, 700 µL of the supernatant were transferred into a new 2 mL tube, 700 µL chloroform/isoamyl alcohol were added, the mixture was shaken, incubated on ice for 10 min, and centrifuged at 4,600 × g for 10 min at room temperature. DNA was precipitated by transferring 600 µL of the supernatant into a new 1.5 mL tube containing 200 µL PEG 6,000 (30% (w/v)) and 100 µL 5 M NaCl. The mixture was incubated at room temperature for 20 min and centrifuged at 20,240 × g at room temperature for 15 min to pellet the DNA. DNA pellets were washed with 70% (v/v) EtOH twice and dried using a vacuum centrifuge. Dried pellets were re-dissolved in 50 µL 1 × TE buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid (EDTA); adjusted to pH 8.0 with HCl) and incubated at 42 °C for 2 h to facilitate re-dissolving. Extracted DNA was inspected using gel electrophoresis on 1% (w/v) agarose gels stained with SYBR Green Solution I (Thermo Fisher Scientific, Waltham, MA, USA).
Real-time PCR
Real-time PCR assays for the quantification of total bacteria, total fungi, Basidiomycota, Ascomycota, Acidobacteria, Actinobacteria, Alpha-, Beta-, Gammaproteobacteria, Bacteriodetes, and Firmicutes were performed in a Peqstar 96Q thermocycler (PEQLAB, Erlangen, Germany). The composition of the mastermix and the thermocycling conditions correspond to those described by Beule et al. (2020). Total archaea were amplified using the primer pair ARC787F and ARC1059R (Yu et al. 2005). The reaction mixture comprised of 3 µL mastermix (double distilled H2O, buffer (10 mM Tris–HCl, 50 mM KCl, 2.0 mM MgCl2, pH 8.3 at 25 °C); 100 μM of each deoxynucleoside triphosphate (New England Biolabs, Beverly, Massachusetts, USA); 0.3 μM of each primer; 0.1 × SYBR Green I solution (Thermo Fisher Scientific, Waltham, MA, USA); 1 μg μL−1 bovine serum albumin; 0.03 u μL−1 Hot Start Taq DNA Polymerase (New England Biolabs, Beverly, Massachusetts, USA)) and 1 μL of template DNA or double distilled H2O for a negative control. Thermocycling conditions were as follows. Initial denaturation at 95 °C for 2 min followed by 40 cycles of denaturation (95 °C for 20 s), annealing (62 °C for 30 s), and elongation (68 °C for 20 s). Final elongation was performed at 68 °C for 5 min. Following amplification, melting curves were generated by step-wise heating of the PCR product from 65 to 95 °C by 0.5 °C per step under continuous fluorescence measurement. DNA extracts were tested for PCR inhibitors according to Guerra et al. (2020) and diluted 1:50 (v/v) in double distilled H2O prior to PCR to overcome PCR inhibition. Real-time PCR data have been deposited at the BonaRes repository (https://doi.org/10.20387/bonares-y3je-zz30).
Library preparation and amplicon sequencing of soil fungi
Sequencing libraries were prepared by amplifying the fungal ITS1 region using the primer pair ITS1-F_KYO2 (5’-TAGAGGAAGTAAAAGTCGTAA-3’) (Toju et al. 2012) / ITS86R (5’-TTCAAAGATTCGATGATTCA-3’) (Vancov & Keen 2009). PCR reactions were carried out in 96-well plates in an Eppendorf Mastercycler EP Gradient S thermocycler (Eppendorf, Hamburg, Germany) in 25 µL reaction volume within one PCR run using the same mastermix for all libraries. The reaction volume comprised 18.75 µL mastermix and 6.25 µL template DNA diluted 1:50 (v/v) in double distilled H2O or double distilled H2O for negative a control. The mastermix contained double distilled H2O, buffer (10 mM Tris–HCl, 50 mM KCl, 2.0 mM MgCl2, pH 8.3 at 25 °C), 100 µM of each deoxynucleoside triphosphate (New England Biolabs, Beverly, Massachusetts, USA), 0.5 µM of each primer (ITS1-F_KYO2 / ITS86R), 1 mg mL−1 bovine serum albumin, and 0.03 u µL−1 Hot Start Taq DNA Polymerase (New England Biolabs, Beverly, Massachusetts, USA). Each primer was a mixture of primer with (50%) and without (50%) Illumina TruSeq adapters (5’-GACGTGTGCTCTTCCGATCT-3’ for the forward primer and 5’-ACACGACGCTCTTCCGATCT-3’ for the reverse primer) at the 5`-end. The thermocycling conditions were as per (Beule and Karlovsky 2021a, b): initial denaturation (95 °C for 2 min), 3 touch-up cycles of denaturation (95 °C for 20 s), annealing (50 °C for 30 s), and elongation (68 °C for 60 s) followed by 25 cycles of denaturation (95 °C for 20 s), annealing (58 °C for 30 s), and elongation (68 °C for 60 s), and final elongation (68 °C for 5 min). Following thermocycling, successfulness of the amplification was confirmed by visualizing 2 µL of the PCR product on 1.7% (w/v) agarose gels stained with SYBR Green I solution (Thermo Fisher Scientific GmbH, Dreieich, Germany). Gel electrophoresis was performed at 4.6 V cm−2 for 60 min. Libraries were shipped to the facilities of LGC Genomics (Berlin, Germany) for a second amplification step using standard i7- and i5- sequencing adapters and Illumina sequencing. The second PCR was performed in 20 µL reaction volume containing 1 × MyTaq buffer (Bioline GmbH, Luckenwalde, Germany), 15 pmol of each forward and reverse i7- and i5- sequencing adapters, 2 µl of BioStabII PCR Enhancer (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), and 0.075 u µL−1 MyTaq DNA polymerase (Bioline GmbH, Luckenwalde, Germany).The thermocycling conditions were as follows: initial denaturation (96 °C for 1 min), 3 touch-up cycles of denaturation (96 °C for 15 s), annealing (50 °C for 30 s), and elongation (68 °C for 90 s) followed by 7 cycles of denaturation (96 °C for 15 s), annealing (58 °C for 30 s), and elongation (68 °C for 90 s), and final elongation (70 °C for 2 min). DNA quantity was assessed on agarose gels and indexed sequencing libraries were pooled. The multiplexed libraries were sequenced using on an Illumina MiSeq using V3 chemistry (2 × 300 bp) (Illumina, Inc., San Diego, CA, USA) at LGC Genomics, Berlin, Germany. Amplicon sequencing data have been deposited at NCBI’s Short Read Archive (PRJNA885015).
Processing of amplicon sequencing data
Raw paired-end sequencing data were demultiplexed using Illumina’s bcl2fast v. 2.20 (Illumina, San Diego, CA, USA) and sorted by their barcodes. Barcodes with more than two mismatches as well as one-sided and conflicting barcode pairs were excluded. Furthermore, Illumina sequencing adapters and primers (allowing three mismatches per primer) were clipped, whereas reads shorter than 100 bp were excluded. Afterwards, reads were processed in QIIME 2 version 2022.2 (Bolyen et al. 2019). Sequence quality was evaluated utilizing the ‘q2-demux’ plugin. Using DADA2 (Callahan et al. 2016), forward and reverse reads were truncated to 220 and 180 bp, respectively, quality filtered (allowing two expected errors per read), and merged. Moreover, chimeras and singletons were removed. Afterwards, reads were clustered into exact amplicon sequence variants (ASVs). ASVs were taxonomically classified against the UNITE database version 8.3 QIIME developer release (Abarenkov et al. 2021) using a scikit-learn Naive Bayes machine-learning classifier (‘q2-fit-classifier-naive-bayes’ and ‘q2- classify-sklearn’ plugin) in the ‘balanced’ configuration ([6,6]; 0.96) as suggested by Bokulich et al. (2018). After non-fungal ASVs were removed, 3,945,026 sequence counts remained. Sequence counts were normalized to 41,564 counts per sample using scaling with ranked subsampling (SRS) (Beule and Karlovsky 2020) in the R environment v. 4.2.1 (R Core Team 2013) utilizing the ‘SRS’ R package version 0.2.3 (Heidrich et al. 2021). SRS curves were generated using the same R package. The normalized dataset contained 2,949 fungal ASVs.
Soil properties
Soil pH was determined from sieved (< 2 mm) and air-dried soil in deionized H2O at a ratio of 1:2.5 (soil/water (w/v)). Soil bulk density was determined from 250-cm3 soil cores dried at 105 °C for 24 h. WFPS was determined from the same sample assuming a particle density of 2.65 g cm−3. Double lactate-extractable phosphorus (PDL) and potassium (KDL) were determined from sieved (< 2 mm) and air-dried soil as per (VDLUFA 1991a). Calcium chloride-extractable magnesium (MgCaCl2) was determined as described previously (VDLUFA 1991b). Carbonates in sieved (< 2 mm) and air-dried soil were removed using acid fumigation as per Harris et al. (2001) and concentrations of SOC and total nitrogen were determined on a CNS elemental analyzer (Vario EL Cube, Elementar, Germany). Soil properties data have been deposited at the BonaRes repository (https://doi.org/10.20387/bonares-y3je-zz30).
Statistical analysis
For each tree species, differences in soil properties, microbial abundance, and earthworm parameters among the different sampling locations within the agroforestry system (i.e. tree row, 1 m, 7 m, and 18 m into the crop row) and the monoculture cropland were tested using one-way analysis of variance (ANOVA) followed by Tukey honestly significant difference (HSD). Relationships between parameters (i.e. soil properties, microbial abundance, and earthworm parameters) were explored using Spearman’s rank correlation test. For the soil fungal community, alpha (Shannon diversity, Chao1 index, and Pielou’s evenness) and beta diversity (Bray–Curtis dissimilarity index) were computed using ‘vegan’ R package v. 2.6.2 (Oksanen et al. 2013). Differences in fungal community composition among the different sampling locations within the agroforestry system and the monoculture cropland were identified using permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis dissimilarities using the ‘vegan’ R package. Multivariate homogeneity of dispersions was tested using the same R package. Furthermore, differential abundance analysis using data collapsed at genus level was performed using the ‘metacoder’ R package v. 0.3.5 (Foster et al. 2017). The same package was utilized to generate heat trees. For each tree species, differences in alpha diversity and taxonomic groups of fungi among the different sampling locations within the agroforestry system and the monoculture cropland were tested using one-way ANOVA followed by Tukey HSD test or Kruskal–Wallis test followed by Dunn’s test. All data were tested for normality of residuals (Shapiro–Wilk test) and homogeneity of variance (Levene’s test). Statistical significance was considered at p < 0.05. All tests were performed in R version 4.1.2.
Results
Mean earthworm density in the tree rows of the agroforestry system was 2.9 to 12.3 times greater than in the crop rows and the monoculture cropland (Fig. 2A) (p ≤ 0.0216). Likewise, mean earthworm biomass was 5.7 to 34.8 times greater in the tree rows than in the crop rows and the monoculture cropland (Fig. 2B) (p ≤ 0.0014). A gradual decline in earthworm density from the trees into the crop rows and monoculture cropland was evident for all three tree species (Fig. 2A). For example, earthworm density was greater at 1 m than at 18 m distance from the trees and monoculture cropland under poplar Max 1 cultivation (Fig. 2A) (p = 0.0344). The gradual decline in population density was mainly driven by endogeic (Figure S2 A) rather than anecic species (Figure S2 C). Across sampling locations, population density and biomass of earthworms were strongly positively correlated (r = 0.84; p < 0.0001). In total, five different earthworm species of three different ecological groups were found at our study site: Allolobophora chlorotica, Aporrectodea caliginosa, Aporrectodea rosea (endogeic species), Lumbricus rubellus (epigeic species), and Lumbricus terrestris (anecic species). The monoculture cropland was dominated by endogeic species with no anecic and epigeic species present (Fig. 3). Anecic species were always present in the tree rows of all three tree species ranging from 13.8 to 44.9% of the total community and were occasionally recovered in the crop rows (Fig. 3). In the poplar Max 1 system, the epigeic species (i.e. L. rubellus) was found in half of all replicate plots in the tree row and at 1 m distance from the trees as well as in a quarter of all the replicate plots at 7 m distance (Fig. 3). Epigeic earthworms were only detected in one replicate plot of the tree rows in the black locust system (Fig. 3).
Earthworm density (A) and biomass (B) in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
Relative abundance of ecological groups of earthworms in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars represent individual samples (n = 4)
We quantified 12 groups of soil microorganisms (total bacteria, total fungi, total archaea, Basidiomycota, Ascomycota, Acidobacteria, Actinobacteria, Alpha-, Beta-, Gammaproteobacteria, Bacteriodetes and Firmicutes) and found only positive or no effects of agroforestry on microbial abundance. Total soil bacterial abundance was greater in the tree rows than at 18 m distance from the trees within the crop rows (Fig. 4A) (p ≤ 0.0378). In the poplar Max 1 system, gene abundance of bacteria as well as Actinobacteria was greater under the trees as compared to 7 and 18 m into the crop row and the monoculture cropland (Fig. 4A, C) (p ≤ 0.0288). In the same system, greater abundance of Acidobacteria and Bacteroidetes was detected in the tree row as compared to the crop row and the monoculture cropland (Fig. 4B, S3 A) (p ≤ 0.0290) and Firmicutes were more abundant under the trees than at 18 m distance into the crop row and the monoculture cropland (Figure S3 B) (p ≤ 0.0092). The abundance of Basidiomycota in the poplar Max 1 system was greater in the tree row than in the crop row at 7 and 18 m distance and in the monoculture cropland (Figure S4 C) (p ≤ 0.0313). Gene abundances of total soil fungi, Ascomycota, total archaea, Alpha-, Beta-, and Gammaproteobacteria did not differ among sampling locations (Figure S4 A-B, S5, S6).
16S rRNA gene abundance of total bacteria (A), Acidobacteria (B), and Actinobacteria (C) in soil of a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
The soil fungal community was dominated by Ascomycota (51.4 ± 16.7%), Basidiomycota (32.4 ± 22.4%), and Mortierellomycota (5.8 ± 4.0%). The dominant fungal classes were Sordariomycetes (28.7 ± 14.4%), Agaricomycetes (27.4 ± 22.8%), Dothideomycetes (6.9 ± 5.3%), and Mortierellomycetes (5.8 ± 4.0%) (Fig. 5A). Acremonium (8.6 ± 6.2%), Mortierella (4.9 ± 3.4%), Coprinellus (4.5 ± 8.9%), Laccaria (3.2 ± 9.8%), and Agrocybe (2.9 ± 10.7%) were the most abundant genera in the dataset. Alpha diversity (Shannon diversity, Chao1 index, and Pielou’s evenness) did not differ among sampling locations (i.e. tree row, 1, 7, 18 m crop rows, and monoculture cropland) within each agroforestry system (i.e. poplar Fritzi Pauley, poplar Max 1, and black locust agroforestry system) (Table S1). Sampling location significantly affected fungal community composition (Table S2) (p = 0.0001) and was driven by the differences between the tree row and the arable land (crop row and monoculture cropland) (Fig. 5B). Multivariate homogeneity of dispersions was given under all PERMANOVA test conditions performed (p ≥ 0.32). Differential abundance analysis was visualized by heat trees (Figure S 7) and a total of 15 genera and species were identified whose relative abundance was either positively or negatively affected by agroforestry (Fig. 6). Relative abundance of Gamsia, Ilyonectria, Laccaria, and Preussia spp. were generally enhanced by the tree rows, whereas the increased relative abundance of Inocybe spp. was specific for poplar Max 1 tree rows (Fig. 6). Relative abundance of Protomyces spp., Sporobolomyces spp., and Blumeria graminis showed a positive trend with increasing distance from the trees. Among these, Blumeria graminis was absent in the tree rows. Relative abundance of Mycosphaerella tassiana as well as Dioszegia, Itersonilia, Lectera, Leucosporidium, Microdochium, and Neosetophoma spp. was overall higher in the crop row or monoculture cropland compared to the tree rows; however, no trends regarding sampling distance were observed (Fig. 6).
Soil fungal community composition in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, soil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row (n = 4). Relative abundances of dominant (≥ 0.5% mean relative abundance) fungal classes (A) are shown. Beta diversity is shown by non-metric multidimensional scaling (NMDS) of Bray–Curtis dissimilarities (B). Circles, squares, and triangles represent individual data points. AF = agroforestry
Differentially abundant genera and species in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, soil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row (n = 4)
In the black locust system, SOC and total N concentrations in topsoil (upper 0–5 cm soil) were 54 to 97% greater in the tree rows than in the crop row and monoculture cropland (Fig. 7) (p ≤ 0,0004). Likewise, in the poplar Max 1 system, SOC concentrations in topsoil were greater in the tree rows than in the crop row and monoculture cropland (Fig. 7A) (p ≤ 0.0376). Tree rows of poplar Fritzi Pauley showed greater SOC concentrations than the crop row at 18 m distance (Fig. 7A) (p = 0.0093) but similar total N concentrations among all sampling locations (Fig. 7B). Soil bulk density showed slight differences across sampling locations, however, no distinct spatial pattern among sampling locations was detected (Table S3). In the black locust system, MgCaCl2 was greater under the trees than in the crop row (p = 0.0001 – 0.0004) (Table S3). Similarly, MgCaCl2 in the system with poplar Fritzi Pauley was greater in the tree row than in the crop row and monoculture cropland (p = 0.0006 – 0.0086) (Table S3). In both, the poplar Max 1 and the black locust system, KDL increased in the tree row as compared to the crop row and monoculture cropland (p < 0.0001 – 0.0073) (Table S3). Tree rows of poplar Max 1 showed greater PDL as compared to monoculture cropland (p = 0.0106). Earthworm density and biomass were positively correlated to SOC and total N concentrations, KDL, MgCaCl2, and WFPS (r = 0.35 – 0.56; p < 0.0001 – 0.011) but not PDL (r = 0.04 – 0.14; p = 0.32 – 0.77) (Figure S8). Positive relationships were found between soil bacteria, Bacteroidetes, Acidobacteria, and Actinobacteria and SOC and total N concentrations, KDL, MgCaCl2, and PDL (r = 0.28 – 0.54; p < 0.0001 – 0.041) (Figure S9).
Soil organic carbon (SOC) (A) and total N concentrations (B) in 0–5 cm topsoil in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
Discussion
Improved soil fertility through agroforestry
In agreement with previous findings (e.g. Pardon et al. 2017), soil fertility (i.e. SOC, total N, and extractable nutrients (KDL and MgCaCl2)) increased under the trees (Fig. 7, Table S3). This increase was likely due to the input of above- and belowground tree litter-derived nutrients, which declines with increasing distance from the trees (Schmidt et al. 2021). Although tree-derived nutrient inputs can reach several meters into the crop row (Schmidt et al. 2021), increased soil fertility did not gradually extend into the crop rows which we attribute to the relatively young age (12 years) of our system (cf. Pardon et al. 2017). We expect that as our system ages and more tree litter is deposited, the increase in soil fertility under the trees will increasingly extend into the crop rows.
Agroforestry promotes earthworm communities
In the present study, earthworms were sampled using AITC extraction without hand sorting. Compared to hand sorting combined with AITC extraction (e.g. as per ISO (2018) 23611–1), using exclusively AITC extraction for earthworm sampling may result in reduced efficacy for juveniles and endogeic species whereas adult anecic earthworms are well recovered (e.g.Pelosi et al. 2009; Čoja et al. 2008). Hand sorting, however, is rarely feasible for large-scale studies as it requires a substantial amount of labour and may prolong the sampling campaign (Iannone et al. 2012). Furthermore, if additional on-site data needs to be collected, chemical extraction is preferred over hand sorting since it does not physically disturb the soil (Iannone et al. 2012; Lees et al. 2016; Tóth et al. 2020).
Unlike Price and Gordon (1998), we did not detect tree-species effects on earthworm abundance, diversity or community composition at our study site. In line with previous studies (Cardinael et al. 2019; D'Hervilly et al. 2022), population size of earthworms was greater in the tree rows as compared to the crop rows (Fig. 2A). In agreement with the results of Cardinael et al. (2019), the increase in density was evident for all three ecological groups (Figure S2). Furthermore, we were able to demonstrate that earthworm abundance decreases with increasing distance from the trees (Fig. 2A). This gradual decline was driven mainly by endogeic species (cf. Figure S2 A, C, E), which almost exclusively colonize the topsoil. A recent study conducted in two similarly managed alley-cropping agroforestry systems of similar age in Germany found that tree roots in the topsoil of the tilled crop rows are scarce (Schmidt et al. 2021). Consequently, endogeic species in the crop row likely benefited from tree litter input in vicinity of the trees rather than tree root litter. To our best knowledge, the present study is the first to investigate earthworm communities at more than two distances from the tree rows into the crop rows. Although a greater number of sampling locations is demanding, we believe that such study designs are essential to improve the characterization of spatial gradients within agroforestry systems, and thus, the extent by which the tree rows influence the crop rows and vice versa.
Earthworm communities in the tree rows were characterized by a shift towards anecic earthworms (Fig. 3), which is in agreement with previous findings (D'Hervilly et al. 2020, 2021). In contrast to the gradual decrease of endogeic earthworms with increasing distance from the trees, density of anecic earthworms did not decline gradually but rapidly dropped which is most likely due to the tillage in the crop rows. It is well established that anecic earthworms benefit from reduced tillage (Chan 2001; Ernst & Emmerling 2009). Unsurprisingly, the absence of tillage under the trees has recently been identified as a main driver for the abundance increase of anecic earthworms in the tree rows of agroforestry systems (Cardinael et al. 2019). Since anecic earthworms feed on the soil surface, litter fall and understory vegetation should also be considered as promoting factors for deep burrowing species. Additionally, Gilbert et al. (2014) were able to show that saprophagous earthworms, including Lumbricus terrestris, benefit from tree fine roots and fine root associated-mycorrhizae as a feeding source. In order to disentangle the impact of soil management (i.e. tillage) and food resource availability (i.e. litter, fine roots, mycorrhizae, and understory vegetation) on the abundance of anecic earthworms, studies in no-till agroforestry systems are required.
The sporadic detection of epigeic individuals identified as L. rubellus under as well as in close proximity to the trees (Figure S2 E, F) is in accordance with the results of D'Hervilly et al. (2020). We argue that this is likely due to tree litter input, as those species require a permanent surface cover with organic material and are therefore mostly absent in arable croplands. We speculate that depending on the size of the agroforestry system, the colonization of tree rows by epigeic earthworms requires several years to reach a spatially homogeneous equilibrium state.
Effects of agroforestry on soil microbial population size
The population size of total bacteria, Acidobacteria, and Actinobacteria was greater in the vicinity of trees in all agroforestry systems (Fig. 4), which is in line with results of recent studies (Beule et al. 2020; Guillot et al. 2021). The positive effect of trees in agroforestry systems on the abundance of soil microorganisms is well-described (Banerjee et al. 2016; Beuschel et al. 2019; Beule et al. 2020; Guillot et al. 2021; Luo et al. 2022) and is likely due to differences in soil management (i.e. absence of tillage in the tree rows versus tilled crop rows) and vegetation cover (i.e. woody perennials versus annual crops) (Beule et al. 2022a). Furthermore, some studies were able to demonstrate that microbial abundance increases with decreasing distance from the trees (e.g. Guillot et al. 2019, Beule et al. 2020; D'Hervilly et al. 2021; Luo et al. 2022). At our study site, no such trends were observed except for total bacteria (Fig. 4A), Actinobacteria (Fig. 4C), Firmicutes (Figure SI2 B), and Bacteriodetes (Figure SI 3C) in the poplar Max 1 system. In contrast to previous studies where trees were either not harvested at all (Guillot et al. 2019, 2021; D'Hervilly et al. 2021) or harvested four to five years (Beule et al. 2020; Luo et al. 2022) prior to soil sampling, the aboveground biomass of the trees at our study site was harvested one year prior to soil sampling. Therefore, we suggest that tree harvesting temporally alters soil microbial communities throughout agroforestry systems. We relate these alterations to changes in substrate input due to reduced tree litter input and/or altered tree root functioning (e.g. dying off of roots, changes in quantity and quality of root exudation). Furthermore, this hypothesis may explain why in contrast to previous studies (Beuschel et al. 2019; Beule et al. 2020; Guillot et al. 2021), soil fungal abundance at our study site was not promoted by agroforestry (Figure SI 3). Further studies exploring temporal dynamics along tree rotation cycles are required to investigate this hypothesis.
Agroforestry alters the soil mycobiome and putative phytopathogen abundance
Our finding that agroforestry does not affect alpha (Table S1) but beta diversity of soil fungi (Table S2, Fig. 5B) agrees with previous studies on the soil mycobiome of agroforestry systems (Beule et al. 2021, Beule & Karlovsky 2021b). Differences in fungal community composition were mainly driven by the tree rows (Fig. 5B) which are known to exert strong influence on the soil fungal community even few months after tree planting (Beule & Karlovsky 2021b). Across all three tree species, the tree-row associated mycobiome was characterized by the promotion of genera harbouring ectomycorrhizal fungi (EMF) such as Laccaria and Preussia spp. (Fig. 6). Notably, affiliates of the genus Inocybe, harbouring EMF that can associate with poplar (e.g. Long et al. 2016), were strongly promoted under poplar Max 1 (Fig. 6) which indicates a tree species-specific tree-EMF interaction. Colonization of roots by mycorrhiza is often advantageous for plant growth as it improves nutrient and water acquisition. Under field conditions, poplar and black locust trees can associate with both arbuscular (AMF) as well as ectomycorrhizal fungi (e.g. Khasa et al. 2002; Bratek et al. 1996). For poplar trees, colonization rates of AMF and EMF have been shown to be poplar genotype-specific (Khasa et al. 2002), whereas black locust genotype specificity of mycorrhizal associations has not been investigated yet. Our findings on EMF highlight the importance of investigating several tree species (e.g. hardwood and fast-growing tree species) at the same study site in order to disentangle tree-species from study-site effects.
Concerns regarding an increased risk of crop diseases is among the main impediments of farmers to implement temperate agroforestry. In our study, a gradual decrease in relative abundance of Blumeria graminis (formerly known as Erysiphe graminis) with decreasing distance from tree rows was observed (Fig. 6). B. graminis is the causal agent of powdery mildew, one of the most common cereal diseases worldwide that can reduce grain quality and cause significant yield losses in temperate zones (e.g. Bélanger et al. 2002; Dreiseitl 2011; Dean et al. 2012). The ability of B. graminis to cause disease is host specific and thus forma specialis dependent (Wyand and Brown 2003). Sequencing of short-read amplicons (2 × 300 bp) did not enable identification of B. graminis on forma specialis level, which is not surprising considering the limited taxonomic resolution of short-read amplicon sequencing (Heidrich & Beule 2022). Furthermore, B. graminis can overwinter in form of cleistothecia on plant residues (Zhang et al. 2005). Thus, it was not possible to relate B. graminis to any of the host crops that were grown in the crop rotation at our study site. Hence, we refer to B. graminis as a putative pathogen. Still, our results provide the first evidence of a positive effect of tree rows in temperate agroforestry systems on the reduction of the putative pathogen B. graminis. As microbial antagonists of B. graminis have been isolated in Germany (Köhl et al. 2019), antagonistic interactions in soil and on aboveground crop parts may have contributed to the suppression of B. graminis at our study site. In addition to microbial antagonism, the ability of earthworms to suppress soil-born fungal phytopathogens is well recognized (e.g. Plaas et al. 2019; Stephens et al. 1994). Thus, the gradual increase in endogeic earthworm abundance with decreasing distance from the trees may have enhanced biological control within the crop rows and thereby contributed to the suppression of B. graminis.
In addition to potential biological control, tree rows in agroforestry systems increase structural diversity and act as physical barriers which is expected to lower the spread of crop diseases through the dilution of the host crop (host dilution effect) (Beule et al. 2019b). In 2019, Kanzler et al. reported reduced wind speed and evaporation rates at our study site under the agroforestry as compared to the monoculture cropland. Furthermore, reductions of wind speed and evaporation were dependent on the distance from the trees (Kanzler et al. 2019). Such microclimatic alterations are known to affect the epidemiology of plant diseases (e.g. Aust & von Hoyningen-Huene 1986, Waggoner 1965). Overall, the enhanced control of B. graminis in the agroforestry as compared to the monoculture cropland system cannot be attributed directly to one of the factors listed above. We rather expect that the control of B. graminis was due to a combination of factors that cannot be disentangled in our experimental setting.
As of writing, the study conducted by Beule et al. (2019b) is the only study that investigated the effect of temperate agroforestry on crop health. Their results revealed that colonization of oilseed rape plants with Verticillium longisporum and wheat grain with Fusarium tricinctum was lower in temperate agroforestry systems compared to monoculture cropland systems. Colonization of wheat and barley grain and oilseed rape plants with other major fungal pathogens did not differ between agroforestry systems and monoculture systems (Beule et al. 2019b). Furthermore, the authors observed a relationship between abundance of a phytopathogen and distance to the trees only for the phytopathogen Leptosphaeria biglobosa in oilseed rape plants. Considering our findings on B. graminis, we hypothesize that suppression of fungal phtyopathogens within agroforestry systems is limited to certain pathogens and is a function of the distance to the trees (i.e. suppressiveness increases as the distance to the trees decreases). Studies on diseases and disease related factors in temperate agroforestry systems are scarce. We suggest that future research should investigate crop diseases in agroforestry systems of different spatial designs (e.g. wide versus narrow crop allays) as well as management practices. New findings could help farmers to optimize the design of future agroforestry systems in order to maximize the beneficial effects of these systems on disease control.
Conclusion
The integration of tree rows into arable land (agroforestry) increased the abundance of soil bacteria and earthworms (anecic, endogeic, and epigeic species) as compared to monoculture cropland. We attribute this mainly to the absence of tillage and the input of large amounts of tree litter under the trees. Community composition of soil fungi was altered by the tree rows, resulting in a tree-row associated mycobiome, which was particularly characterized by an increased proportion of EMF. The tree-row associated mycobiome not just enhances overall fungal diversity of agroforestry systems but is also expected to alter soil functions such as nutrient cycling. As the distance from the trees decreased, the proportion of Blumeria graminis, the causal agent of powdery mildew, decreased. We suggest that enhanced microbial antagonism, increased earthworm densities and/or altered microclimatic conditions contributed to the suppression of B. graminis within the agroforestry system. Whereas distinct tree-distance effects were observed, tree-species effects were identified as a minor driver of the abundance and composition of soil communities at our study site. Overall, agroforestry benefits the abundance, diversity, and function of soil biota and may enhance soil suppressiveness. Future research should investigate crop diseases in agroforestry systems of different spatial designs and management practices in order to maximize the beneficial effects of these systems on disease control.
References
Abarenkov K, Zirk A, Piirmann T, Pöhönen R, Ivanov F, Nilsson RH, Kõljalg U (2021) UNITE QIIME release for eukaryotes. Version 10.05.2021. UNITE Community. https://doi.org/10.15156/BIO/1264819
Allen SC, Jose S, Nair PKR, Brecke BJ, Nkedi-Kizza P, Ramsey CL (2004) Safety-net role of tree roots: evidence from a pecan (Carya illinoensis K. Koch)–cotton (Gossypium hirsutum L.) alley cropping system in the southern United States. For Ecol Manage 192:395–407. https://doi.org/10.1016/j.foreco.2004.02.009
Aponte C, García LV, Marañón T (2013) Tree species effects on nutrient cycling and soil biota: A feedback mechanism favouring species coexistence. For Ecol Manage 309:36–46. https://doi.org/10.1016/j.foreco.2013.05.035
Aust H, Hoyningen-Huene JV (1986) Microclimate in relation to epidemics of powdery mildew. Annu Rev Phytopathol 24:491–510. https://doi.org/10.1146/annurev.py.24.090186.002423
Banerjee S, Baah-Acheamfour M, Carlyle CN, Bissett A, Richardson AE, Siddique T, Bork EW, Chang SX (2016) Determinants of bacterial communities in Canadian agroforestry systems. Environ Microbiol 18:1805–1816. https://doi.org/10.1111/1462-2920.12986
Bélanger RR, Bushnell WR, Dik AJ, Carver TL (2002) The powdery mildews: a comprehensive treatise. American Phytopathological Society (APS Press).
Beule L, Karlovsky P (2020) Improved normalization of species count data in ecology by scaling with ranked subsampling (SRS): application to microbial communities. PeerJ 8:e9593. https://doi.org/10.7717/peerj.9593
Beule L, Karlovsky P (2021a) Tree rows in temperate agroforestry croplands alter the composition of soil bacterial communities. PloS one 16:e0246919. https://doi.org/10.1371/journal.pone.0246919
Beule L, Karlovsky P (2021b) Early response of soil fungal communities to the conversion of monoculture cropland to a temperate agroforestry system. PeerJ 9:e12236. https://doi.org/10.7717/peerj.12236
Beule L, Corre MD, Schmidt M, Göbel L, Veldkamp E, Karlovsky P (2019) Conversion of monoculture cropland and open grassland to agroforestry alters the abundance of soil bacteria, fungi and soil-N-cycling genes. PloS one 14:e0218779. https://doi.org/10.1371/journal.pone.0218779
Beule L, Lehtsaar E, Rathgeb A, Karlovsky P (2019b) Crop diseases and mycotoxin accumulation in temperate agroforestry systems. Sustainability 11:2925. https://doi.org/10.3390/su11102925
Beule L, Lehtsaar E, Corre MD, Schmidt M, Veldkamp E, Karlovsky P (2020) Poplar rows in temperate agroforestry croplands promote bacteria, fungi, and denitrification genes in soils. Front Microbiol 10:3108. https://doi.org/10.3389/fmicb.2019.03108
Beule L, Arndt M, Karlovsky P (2021) Relative Abundances of Species or Sequence Variants Can Be Misleading: Soil Fungal Communities as an Example. Microorganisms 9:589. https://doi.org/10.3390/microorganisms9030589
Beule L, Guerra V, Lehtsaar E, Vaupel A (2022) Digging deeper: microbial communities in subsoil are strongly promoted by trees in temperate agroforestry systems. Plant Soil. https://doi.org/10.1007/s11104-022-05591-2
Beule L, Vaupel A, Moran-Rodas VE (2022b) Abundance, Diversity, and Function of Soil Microorganisms in Temperate Alley-Cropping Agroforestry Systems: A Review. Microorganisms 10:616. https://doi.org/10.3390/microorganisms10030616
Beuschel R, Piepho H-P, Joergensen RG, Wachendorf C (2019) Similar spatial patterns of soil quality indicators in three poplar-based silvo-arable alley cropping systems in Germany. Biol Fertil Soils 55:1–14. https://doi.org/10.1007/s00374-018-1324-3
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. https://doi.org/10.1186/s40168-018-0470-z
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Borden KA, Thomas SC, Isaac ME (2017) Interspecific variation of tree root architecture in a temperate agroforestry system characterized using ground-penetrating radar. Plant Soil 410:323–334. https://doi.org/10.1007/s11104-016-3015-x
Bouché M (1972) Lombriciens de France. Ecologie et Systématique. INRA, Paris
Bratek Z, Jakucs E, Bóka K, Szedlay G (1996) Mycorrhizae between black locust (Robinia pseudoacacia) and Terfezia terfezioides. Mycorrhiza 6:271–274. https://doi.org/10.1007/s005720050136
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Cardinael R, Hoeffner K, Chenu C, Chevallier T, Béral C, Dewisme A, Cluzeau D (2019) Spatial variation of earthworm communities and soil organic carbon in temperate agroforestry. Biol Fertil Soils 55:171–183. https://doi.org/10.1007/s00374-018-1332-3
Chan KY (2001) An overview of some tillage impacts on earthworm population abundance and diversity — implications for functioning in soils. Soil and Tillage Research 57:179–191. https://doi.org/10.1016/S0167-1987(00)00173-2
Čoja T, Zehetner K, Bruckner A, Watzinger A, Meyer E (2008) Efficacy and side effects of five sampling methods for soil earthworms (Annelida, Lumbricidae). Ecotoxicol Environ Saf 71:552–565. https://doi.org/10.1016/j.ecoenv.2007.08.002
Das DK, Chaturvedi OP (2008) Root biomass and distribution of five agroforestry tree species. Agrofor Syst 74:223. https://doi.org/10.1007/s10457-008-9159-9
Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430. https://doi.org/10.1111/j.1364-3703.2011.00783.x
D’Hervilly C, Marsden C, Hedde M, Bertrand I (2020) Sown understory vegetation strips impact soil chemical fertility, associated microorganisms and macro-invertebrates in two temperate alley cropping systems. Agrofor Syst 94:1851–1864. https://doi.org/10.1007/s10457-020-00501-w
D’Hervilly C, Marsden C, Capowiez Y, Béral C, Delapré-Cosset L, Bertrand I (2021) Trees and herbaceous vegetation strips both contribute to changes in soil fertility and soil organism communities in an agroforestry system. Plant Soil 463:537–553. https://doi.org/10.1007/s11104-021-04932-x
D’Hervilly C, Bertrand I, Capowiez Y, Béral C, Delapré-Cosset L, Marsden C (2022) Seasonal variations in macrofauna distribution according to the distance from a herbaceous strip in a Mediterranean alley cropping plot. Applied Soil Ecology 170:104309. https://doi.org/10.1016/j.apsoil.2021.104309
Dreiseitl A (2011) Differences in powdery mildew epidemics in spring and winter barley based on 30-year variety trials. Ann Appl Biol 159:49–57. https://doi.org/10.1111/j.1744-7348.2011.00474.x
Ernst G, Emmerling C (2009) Impact of five different tillage systems on soil organic carbon content and the density, biomass, and community composition of earthworms after a ten year period. Eur J Soil Biol 45:247–251. https://doi.org/10.1016/j.ejsobi.2009.02.002
Foster ZSL, Sharpton TJ, Grünwald NJ (2017) Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLOS Comput Biol 13:e1005404. https://doi.org/10.1371/journal.pcbi.1005404
Gilbert KJ, Fahey TJ, Maerz JC, Sherman RE, Bohlen P, Dombroskie JJ, Groffman PM, Yavitt JB (2014) Exploring carbon flow through the root channel in a temperate forest soil food web. Soil Biol Biochem 76:45–52. https://doi.org/10.1016/j.soilbio.2014.05.005
Guerra V, Beule L, Lehtsaar E, Liao H-L, Karlovsky P (2020) Improved protocol for DNA extraction from subsoils using phosphate lysis buffer. Microorganisms 8:532. https://doi.org/10.3390/microorganisms8040532
Guillot E, Hinsinger P, Dufour L, Roy J, Bertrand I (2019) With or without trees: resistance and resilience of soil microbial communities to drought and heat stress in a Mediterranean agroforestry system. Soil Biol Biochem 129:122–135. https://doi.org/10.1016/j.soilbio.2018.11.011
Guillot E, Bertrand I, Rumpel C, Gomez C, Arnal D, Abadie J, Hinsinger P (2021) Spatial heterogeneity of soil quality within a Mediterranean alley cropping agroforestry system: Comparison with a monocropping system. Eur J Soil Biol 105:103330. https://doi.org/10.1016/j.ejsobi.2021.103330
Harris D, Horwáth WR, Van Kessel C (2001) Acid fumigation of soils to remove carbonates prior to total organic carbon or carbon-13 isotopic analysis. Soil Sci Soc Am J 65:1853–1856. https://doi.org/10.2136/sssaj2001.1853
Heidrich V, Karlovsky P, Beule L (2021) ‘SRS’R Package and ‘q2-srs’ QIIME 2 Plugin: normalization of microbiome data using scaling with ranked subsampling (SRS). Appl Sci 11:11473. https://doi.org/10.3390/app112311473
Heidrich V, Beule L (2022) Are short‐read amplicons suitable for the prediction of microbiome functional potential? A critical perspective. iMeta: e38. https://doi.org/10.1002/imt2.38
Iannone BV, Umek LG, Wise DH, Heneghan L (2012) A simple, safe, and effective sampling technique for investigating earthworm communities in woodland soils: implications for citizen science. Nat Areas J 32:283–292. https://doi.org/10.3375/043.032.0305
Isaac ME, Borden KA (2019) Nutrient acquisition strategies in agroforestry systems. Plant Soil 444:1–19. https://doi.org/10.1007/s11104-019-04232-5
ISO (2018) 23611–1 Soil quality — Sampling of soil invertebrates — Part 1: Hand-sorting and extraction of earthworms
Jose S, Gillespie A, Pallardy S (2004) Interspecific interactions in temperate agroforestry. Agrofor Syst 61:237–255. https://doi.org/10.1023/B:AGFO.0000029002.85273.9b
Kanzler M, Böhm C, Mirck J, Schmitt D, Veste M (2019) Microclimate effects on evaporation and winter wheat (Triticum aestivum L.) yield within a temperate agroforestry system. Agrofor Syst 93:1821–1841. https://doi.org/10.1007/s10457-018-0289-4
Khasa PD, Chakravarty P, Robertson A, Thomas BR, Dancik BP (2002) The mycorrhizal status of selected poplar clones introduced in Alberta. Biomass Bioenerg 22:99–104. https://doi.org/10.1016/S0961-9534(01)00072-1
Köhl J, de Geijn HG-v, Haas LG-d, Henken B, Hauschild R, Hilscher U, der Plas CL-v, van den Bosch T, Wikström M (2019) Stepwise screening of candidate antagonists for biological control of Blumeria graminis f sp tritici. Biological Control 136:104008. https://doi.org/10.1016/j.biocontrol.2019.104008
Lees KJ, McKenzie AJ, Newell Price JP, Critchley CN, Rhymer CM, Chambers BJ, Whittingham MJ (2016) The effects of soil compaction mitigation on below-ground fauna: How earthworms respond to mechanical loosening and power harrow cultivation. Agr Ecosyst Environ 232:273–282. https://doi.org/10.1016/j.agee.2016.07.026
Long D, Liu J, Han Q, Wang X, Huang J (2016) Ectomycorrhizal fungal communities associated with Populus simonii and Pinus tabuliformis in the hilly-gully region of the Loess Plateau, China. Sci Rep 6:24336. https://doi.org/10.1038/srep24336
Luo J, Beule L, Shao G, Veldkamp E, Corre MD (2022) Reduced Soil Gross N2O Emission Driven by Substrates Rather Than Denitrification Gene Abundance in Cropland Agroforestry and Monoculture. J Geophys Res Biogeosci 127:e2021JG006629. https://doi.org/10.1029/2021JG006629
Marsden C, Martin-Chave A, Cortet J, Hedde M, Capowiez Y (2020) How agroforestry systems influence soil fauna and their functions-a review. Plant Soil 453:29–44. https://doi.org/10.1007/s11104-019-04322-4
Mayer S, Wiesmeier M, Sakamoto E, Hübner R, Cardinael R, Kühnel A, Kögel-Knabner I (2022) Soil organic carbon sequestration in temperate agroforestry systems–A meta-analysis. Agr Ecosyst Environ 323:107689. https://doi.org/10.1016/j.agee.2021.107689
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Package ‘vegan’. Community ecology package, version 2: 1-295
Pardon P, Reubens B, Reheul D, Mertens J, De Frenne P, Coussement T, Janssens P, Verheyen K (2017) Trees increase soil organic carbon and nutrient availability in temperate agroforestry systems. Agr Ecosyst Environ 247:98–111. https://doi.org/10.1016/j.agee.2017.06.018
Pelosi C, Bertrand M, Capowiez Y, Boizard H, Roger-Estrade J (2009) Earthworm collection from agricultural fields: Comparisons of selected expellants in presence/absence of hand-sorting. Eur J Soil Biol 45:176–183. https://doi.org/10.1016/j.ejsobi.2008.09.013
Plaas E, Meyer-Wolfarth F, Banse M, Bengtsson J, Bergmann H, Faber J, Potthoff M, Runge T, Schrader S, Taylor A (2019) Towards valuation of biodiversity in agricultural soils: A case for earthworms. Ecol Econ 159:291–300. https://doi.org/10.1016/j.ecolecon.2019.02.003
Price GW, Gordon AM (1998) Spatial and temporal distribution of earthworms in a temperate intercropping system in southern Ontario, Canada. Agrofor Syst 44:141–149. https://doi.org/10.1023/A:1006213603150
R Core Team (2013) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
Schmidt M, Corre MD, Kim B, Morley J, Göbel L, Sharma AS, Setriuc S, Veldkamp E (2021) Nutrient saturation of crop monocultures and agroforestry indicated by nutrient response efficiency. Nutr Cycl Agroecosyst 119:69–82. https://doi.org/10.1007/s10705-020-10113-6
Seiter S, Ingham ER, William RD (1999) Dynamics of soil fungal and bacterial biomass in a temperate climate alley cropping system. Appl Soil Ecol 12:139–147. https://doi.org/10.1016/S0929-1393(98)00163-2
Stephens PM, Davoren CW, Doube BM, Ryder MH (1994) Ability of the lumbricid earthworms Aporrectodea rosea and Aporrectodea trapezoides to reduce the severity of take-all under greenhouse and field conditions. Soil Biol Biochem 26:1291–1297. https://doi.org/10.1016/0038-0717(94)90209-7
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-Coverage ITS Primers for the DNA-Based Identification of Ascomycetes and Basidiomycetes in Environmental Samples. PLOS ONE 7:e40863. https://doi.org/10.1371/journal.pone.0040863
Tóth Z, Szlavecz K, Epp Schmidt DJ, Hornung E, Setälä H, Yesilonis ID, Kotze DJ, Dombos M, Pouyat R, Mishra S, Cilliers S, Yarwood S, Csuzdi C (2020) Earthworm assemblages in urban habitats across biogeographical regions. Appl Soil Ecol 151:103530. https://doi.org/10.1016/j.apsoil.2020.103530
Vancov T, Keen B (2009) Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol Lett 296:91–96. https://doi.org/10.1111/j.1574-6968.2009.01621.x
Varah A, Jones H, Smith J, Potts SG (2013) Enhanced biodiversity and pollination in UK agroforestry systems. J Sci Food Agric 93:2073–2075. https://doi.org/10.1002/jsfa.6148
Varah A, Jones H, Smith J, Potts SG (2020) Temperate agroforestry systems provide greater pollination service than monoculture. Agr Ecosyst Environ 301:107031. https://doi.org/10.1016/j.agee.2020.107031
VDLUFA (1991a) Determination of phosphorus and potassium in the double lactate (DL) extract. VDLUFA method book I, A 6.2.1.2. Darmstadt: VDLUFA-Verlag (in German)
VDLUFA (1991b) Determination of plant-available magnesium in the calcium chloride extract. VDLUFA method book I, A 6.2.4.1. Darmstadt: VDLUFA-Verlag (in German)
Waggoner PE (1965) Microclimate and plant disease. Annu Rev Phytopathol 3:103–126. https://doi.org/10.1146/annurev.py.03.090165.000535
Wang Y, Zhang B, Lin L, Zepp H (2011) Agroforestry system reduces subsurface lateral flow and nitrate loss in Jiangxi Province, China. Agr Ecosyst Environ 140:441–453. https://doi.org/10.1016/j.agee.2011.01.007
WRB-FAO IWG (2015) IUSS Working Group WRB. 2015. World Reference Base for Soil Resources 2014, update 2015 International soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports Nº. 106. FAO Rome
Wyand RA, Brown JKM (2003) Genetic and forma specialis diversity in Blumeria graminis of cereals and its implications for host-pathogen co-evolution. Mol Plant Pathol 4:187–198. https://doi.org/10.1046/j.1364-3703.2003.00167.x
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. https://doi.org/10.1002/bit.20347
Zaborski ER (2003) Allyl isothiocyanate: an alternative chemical expellant for sampling earthworms. Appl Soil Ecol 22:87–95. https://doi.org/10.1016/S0929-1393(02)00106-3
Zhang Z, Henderson C, Perfect E, Carver TLW, Thomas BJ, Skamnioti P, Gurr SJ (2005) Of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol Plant Pathol 6:561–575. https://doi.org/10.1111/j.1364-3703.2005.00303.x
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was supported by the German Federal Ministry of Education and Research (BMBF) in the framework of the Bonares-SIGNAL project (funding codes: 031A562A, 031B0510A, 031B1063A). AV was supported by the joint project MonViA—the German Farmland Biodiversity Monitoring that has been funded by the Federal Ministry of Food and Agriculture (BMEL). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
AV and LB contributed to the conception and design of the study. AV and LB performed the field work. AV, ZB, and LB performed the laboratory work. AV, ZB, and LB performed the statistical analysis. AV, ZB, and LB wrote the first draft of the manuscript. NH, BH, and VEMR contributed resources and critically revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Responsible Editor: Timothy J. Fahey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11104_2023_5932_MOESM1_ESM.jpg
Supplementary file1 (JPG 1994 KB) Figure S1. Allyl isothiocyanate (AITC) extraction of earthworms in the field. Application of 5 L of 0.01 % (w/v) AITC solution to a quarter square meter of soil framed by an open metal frame (A). Surfacing earthworms within the open metal frame following AITC application (B). Photo credit: Anna Vaupel
11104_2023_5932_MOESM2_ESM.pdf
Supplementary file2 (PDF 1128 KB) Figure S2. Density and biomass of endogeic (A, B), anecic (C, D), and epigeic (E, F) earthworms in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
11104_2023_5932_MOESM3_ESM.pdf
Supplementary file3 (PDF 712 KB) Figure S3. 16S rRNA gene abundance of Bacteroidetes (A) and Firmicutes (B) in soil of a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
11104_2023_5932_MOESM4_ESM.pdf
Supplementary file4 (PDF 885 KB) Figure S4. 18S rRNA gene abundance of total fungi (A) and ITS gene abundance of Ascomycota (B), and Basidiomycota (C) in soil of a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
11104_2023_5932_MOESM5_ESM.pdf
Supplementary file5 (PDF 549 KB) Figure S5. 16S rRNA gene abundance of total archaea in soil of a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
11104_2023_5932_MOESM6_ESM.pdf
Supplementary file6 (PDF 875 KB) Figure S6. 16S rRNA gene abundance of Alpha- (A), Beta- (B), and Gammaproteobacteria (C) in soil of a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row. Bars with error bars represent the mean and its standard deviation (n = 4). Dots represent individual data points. Different uppercase letters of the same font indicate statistically significant differences (p < 0.05)
11104_2023_5932_MOESM7_ESM.pdf
Supplementary file7 (PDF 5441 KB) Figure S7. Heat trees visualizing results of differential abundance analysis of soil fungal groups in a paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley (A), poplar Max 1 (B), and black locust (C) are different tree species within the agroforestry system. Within the agroforestry system, soil samples were collected in the tree row as well as at 1 m, 7 m, and 18 m distance from the tree row within the crop row (n = 4). Heat trees were collapsed at genus level. For each panel, a key containing taxonomic labels is provided in the lower left. Within the pairwise heat trees on the upper right, turquoise and brown nodes represent statistically significant (p < 0.05) differences between two sampling locations. Turquoise and brown nodes indicate greater abundance of a taxonomic group in the sampling locations listed in rows and columns, respectively. Different node sizes indicate the number of amplicon sequence variants (ASVs) detected per taxonomic group
11104_2023_5932_MOESM8_ESM.pdf
Supplementary file8 (PDF 315 KB) Figure S8. Relationships between earthworm biomass (A-F), earthworm density (G-L) and soil properties in soil of paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, soil samples were collected in the tree row, at three different distances from the tree row within the crop row as well as in a monoculture cropland (n = 4). Relationships were investigated with Spearman’s rank correlation test (r = Spearman´s rank correlation coefficient). Different colors represent different tree species, whereas different shapes represent different sampling locations.
11104_2023_5932_MOESM9_ESM.pdf
Supplementary file9 (PDF 620 KB) Figure S9. Relationships between soil bacteria (A-E), Bacteroidetes (F-J), Acidobacteria (K-O), and Actinobacteria (P-T) and soil properties in soil of paired temperate alley-cropping agroforestry and monoculture cropland system in Germany. Poplar Fritzi Pauley, poplar Max 1, and black locust are different tree species within the agroforestry system. Within the agroforestry system, topsoil samples were collected in the tree row, at three different distances from the tree row within the crop row as well as in a monoculture cropland (n = 4). Relationships were investigated with Spearman’s rank correlation test (r = Spearman´s rank correlation coefficient). Different colors represent different tree species, whereas different shapes represent different sampling locations
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vaupel, A., Bednar, Z., Herwig, N. et al. Tree-distance and tree-species effects on soil biota in a temperate agroforestry system. Plant Soil 487, 355–372 (2023). https://doi.org/10.1007/s11104-023-05932-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-05932-9